Ultra-deep & UMI error-corrected sequencing seemed to be the best tool for early cancer detection and MRD. Early detection is now moving to methylation (e.g.
GRAIL’s recent CCGA paper) and now MRD is being challenged – this time by 35x WGS sequencing (it’s also being challenged by methylation but that’s for another post).
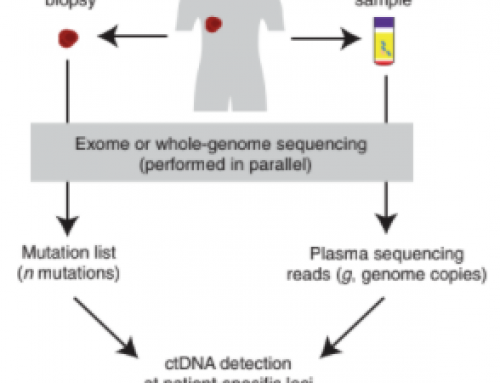
In a paper released on Monday, Dan Landau (WCM, NYGC) and colleagues report on the use of integrated genome-wide mutational analysis with quantitative dynamic monitoring of disease to accurately and sensitively detect cancer at very low levels. The authors claim their with just 1ml plasma and 35x sequencing depth!
Update: C2i Genomics is the spin-out company commercialising the method.
They argue that breadth of sequencing supplants depth to overcome the limitation of low number of DNA fragments in plasma samples…Basically, if you don’t have much ctDNA in your samples, because you have a small amount of sample, the tumour is very smallor the tumour is a “low shedder”, you are unlikely to have any copies of the mutated DNA you’re trying to sequence e.g. EGFR T790M, so it does not matter how great your NGS method is.
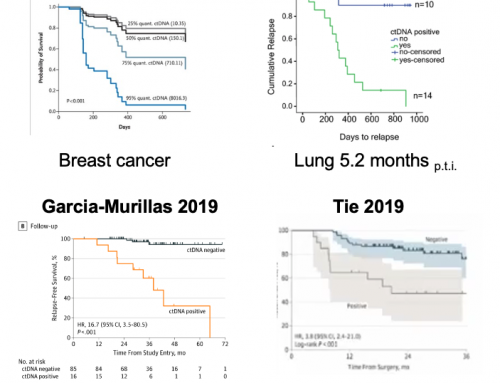
They demonstrate the assay in LUAD; n = 39, CRC; n = 19 and melanoma n = 2 where patients were undergoing surgery (LUAD and CRC) or immunotherapy (melanoma). They confirm that MRDetect tracks tumor responses. The 19 CRC samples used to assess MRD were collected post-surgery (median of 43d) and as with other studies report that patients with detectable ctDNA in their postoperative plasma samples show higher probability for disease recurrence even when there is no other clinical or imaging confirmation at that time.
MRDetect is still a tumor-informed assay i.e. it uses mutation data from sequencing of the tumour biospy or resection to determine what are tumour variants, so has similar logistical challenges to e.g.
Signatera. But because there is no need to design patient specific capture panels the assay has significant advantages over personalised panels. It will be great to see if this is corroborated in head-to-head studies.
Like this:
Like Loading...
Related








[…] Previous […]
[…] whole genome sequencing (WGS) for personalised ctDNA monitoring. Similarly to Zviran et al (see last weeks post) WGS was shown to give ultimate flexibility and the simplest logistics – you just need to […]