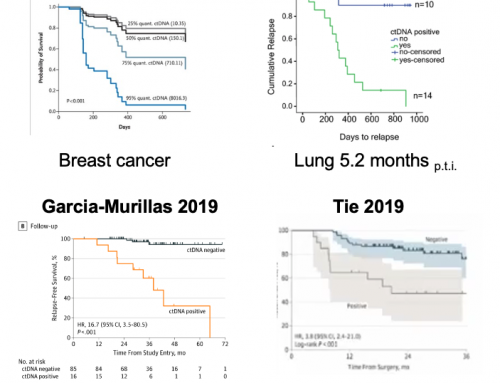
Brain cancer is difficult to diagnose and stratify without an invasive biopsy. Daniel De Carvalho’s group have used their cfChIP-Seq technology to non-invasively diagnose and classify brain tumours, potentially removing the need for surgical tissue biopsies. The power of the assay comes from its use of DNA-methylation profiling rather than mutation calling (as most other liquid biopsies are doing). Epigenetic profiling generates highly specific signatures that have high discriminatory power for classification of different cancers.
In the Nature Medicine paper “Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes” Nassiri et al used a small cohort of 60 patients to derive an initial signature that could be used to detect Gliomas in liquid biopsy. They then expanded this to a larger 160 patient cohort (meningioma, hemangiopericytoma, low-grade glial–neuronal tumors, IDH mutant gliomas, IDH wild-type gliomas and brain metastases from systemic cancers) and were able to determine which of these clinically relevant brain tumour types each patient had. This is a remarkable feat given that current imiaging-based diagnostics can’t distinguish tumours with such similar cell-of-origin lineages.
The power of methylation haplotypes: The assay used just 300 differentially methylated regions (DMRs) and each DMR had a median of eight CpGs that contributed to the signal. Single-loci approaches have been the focus of much development primarily because of the relative ease of development of PCR assays. These can have very high sensitivity and specificity for genetic variants but the subtle changes in methylation can be difficult to pick out. Multiplexing single-locus assays can work but the reality is that signals are not robustly detectable in blood. However methylation haplotyping is becoming standard practice in the assays being developed by Dnamx (the company being spun out of the De Carvalho group) and by many other companies in this space. GRAIL’s recent CCGA paper reported regions of up to 1000bp being used to generate haplotypes. Singlera Genomics have long focused on haplotyping in their Panseer assay. Others include Bluestar Genomics, Cambridge Epigenetix and probably LexentBio (recently acquired by FMI).
Over the last decade many research groups and companies have been developing liquid biopsy to detect circulating tumor DNA (ctDNA) in blood samples (and urine in some cases too). It is likely to be a few years before this cfMeDIP-Seq test is available to clinicians but the future is looking even brighter with the incorporation of epigenomic methods into the liquid biopsy armamentarium.






Leave A Comment