Archive
Categories
-
How do SPRI beads work?April 28th, 2012
-
Index mis-assignment between samples on HiSeq 4000 and X-TenDecember 16th, 2016
-
Cufflinks for Christmas anyone?December 10th, 2012
-
How big would ABI377 plates need to be to deliver NovaSeq X Plus output?October 4th, 2025
-
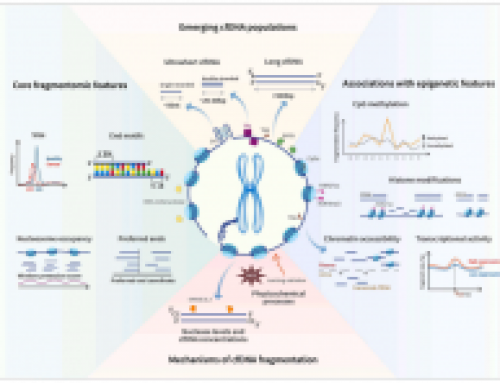
cfDNA fragmentomics reviewedOctober 4th, 2025
-
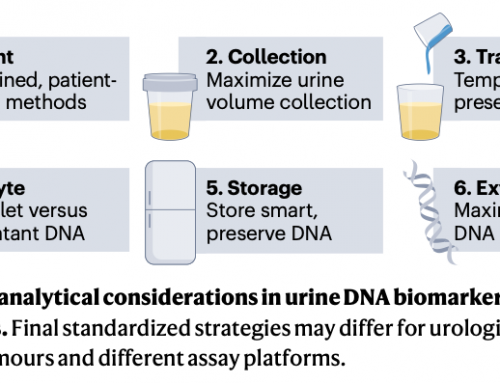
Unlocking the Potential of Urine-Based Liquid BiopsiesOctober 1st, 2025
- No comments have been published yet.
Tags
10X Genomics
16S
CEGX
CoolMPS
CRISPR
ctDNA
diagnostics
DNA methylation
ElementBio
Epigenomics
GeneReader
Hyb&Seq
hydroxymethylation
Illumina
index-swapping
Lego
long-read
methylation
MGI_BGI
MinION
MRD
nanopore
Nanostring
NeoPrep
NGS
NovaSeq
Proteomics
RNA-Seq
single-cell RNA-Seq fingerprinting
single cell
Singular
smMIP







Leave A Comment