Alongside the recent publication from Daniel De Carvalho’s group on glioma detection with cfMeDIP-Seq in Nature Medicine is a report describing the validation of this technology for plasma and urine detection of renal cell carcinoma.
In “Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes” Nuzzo et al in Matthew Freedman’s group at Harvard and The Broad used a cohort of 140 patients to derive a 300 DMR RCC-detection signature. The group generated data in both plasma and urine from RCC patients and reported AUROC of0.979 (plasma) and 0.858 (urine).
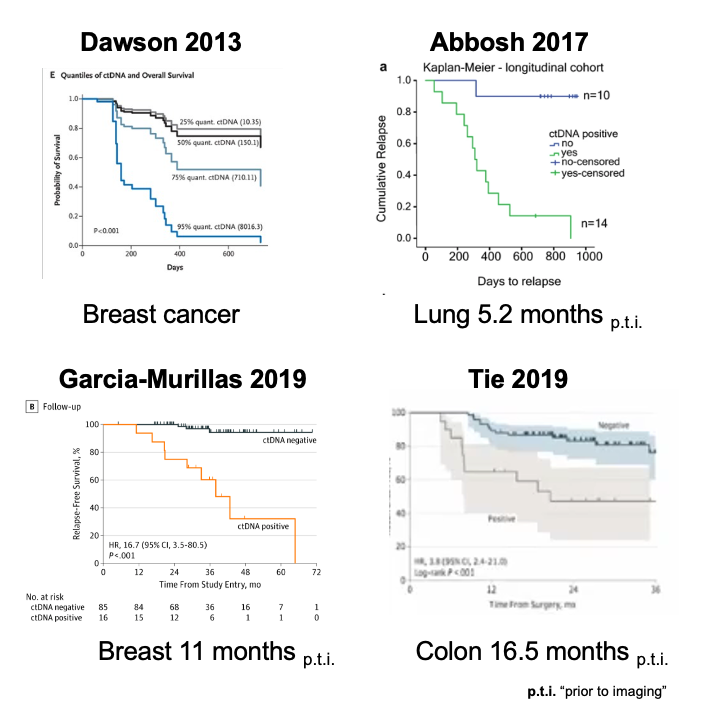
The group highlight previous TCGA data that identified 289 genes with aberrant DNA methylation compared to only 19 recurrently mutated genes, thus underscoring the broad utility of using epigenomic variation alongside or instead of genetic variants.
The paper validates use of cfMeDIP-Seq in liquid biopsy and demonstrates its application to urine. This is likely to be a useful assay for renal cancers where needle biopsies are not favoured.





Leave A Comment