Element Biosciences launched their AVITI sequencing platform (link to spec sheet) today and Illumina now have two new competitors in almost as many months (read my post on Singular from January). The Element pitch is to offer the highest data quality, lowest cost, and best flexibility on a desktop instrument. There have been very few details of the technology, AVIDITY chemistry, or roll-out plans to date, so see below for details on the specs, the apps & data and a brief look at the short-read competitive landscape. I think I can speak for many readers of this blog by saying how exciting this year is starting out!
2022 is going to be a year-to-remember for Genomics afficonado’s. Illumina now has multiple competitors in both the short- and (debatably) long-read space e.g. BGI-MGI, Singular Genomics, PacBio’s Omniome, Genapsys and at least one A.N.Other. And the late 2021 jury verdict, which invalidated Illumina’s “Modified nucleotides” US Patent No. 7,541,444, one of the core Solexa SBS patents that Illumina acquired in 2006, that was set to expire in 2023, means that companies challenging on the SBS chemistry (and Manteia clustering) would be able to enter the market more easily.
Element Biosciences raised approximately $400 million to deliver AVITI and that looks to be money well spent. AVITI is a cheaper instrument that is cheaper to run – what’s not to like?
Element are clearly challenging on cost and this differentiates AVITI from Illumina’s line-up. If you plot output and cost (CapEx or $/Gb) of the Illumina instrument range the various boxes sit on a pretty straight line whereas Element start to plot a new line (here’s hoping for an AVITIxl to compete with NovaSeq). Their fundamentally different chemistry, which uses much less reagent (reducing costs) and generates higher signal to noise (increasing quality) is the star (you’ll understand the pun if you saw the presentation) of the show but lets take a dive into what was launched today and then get on with talking about whether there’s enough to tempt Illumina users and core-labs to make the switch?!

AVITI/AVIDITY overview and specifications
The AVITI launch presentation described the Element Biosciences proprietary AVIDITY chemistry, polony clustering and highlighted the performance, which compares well to NextSeq. This is NOT a carbon-copy of Solexa SBS. Element discussed how challenging it was to build the new system by focusing on only one “Element” so focused on improving everything they could e.g. maximising signal to noise so they could virtually eliminate background to make image analysis as robust as possible.
- Flowcell surface chemistry: Element aimed to deliver the highest possible Contrast-to-Noise (CNR) possible by maximising signal and virtually eliminating background to make image analysis as robust as possible. This required going back to the drawing board to understand what actually drives background noise. They introduce a new “non-stick”, hence low background, and modular surface chemistry to allow alternative primers or macromolecules on the surface – they are obviously thinking more than just DNA sequencing.
- New amplification chemistry – Because the new surface is relatively non-stick it required a modified amplification chemistry that would stick as discrete clusters. Element are using polonies, which appear to be similar to, but not the same as, the tech developed by Mitra/Chruch; and Element clearly don’t want to call their amplification “clustering” or their polonies “clusters”, most likely for marketing or legal reasons.
- New AVIDITY sequencing chemistry – This is a new and proprietary multivalent chemistry that binds the fluorescent chemistry for imaging, the Avidite is then removed so that 3′-blocked nucleotides can be incorporated, and finally the blocking moiety is removed ready for the next sequencing cycle. This chemistry incorporates a “core” of fluorescence that they said helps to amplify or “scale” the signal and because they are not adding a fluorescent moiety to the incorporated bases they get much more flexibility in what can be added to the core – expect to see this as one of their development areas in the future. Additionally, the new chemistry reduces the amount of non-natural nucleotide required, therefore keeps costs-of-goods low, and also enables a larger field-of-view with imaging of smaller pixels, therefore maximising the amount of data that can be captured in an image.
- New PE turnaround – Element introduced a new paired-end chemistry possibly due to patent limitations meaning they could not copy Illumina’s turnaround chemistry – although I don’t know the details on this. NB: as I understand it the PE turnaround chemistry was invented by a Solexa employee who was rewarded with a gift voucher!!!
- New basecalling algorithms – new proprietary algorithms make colour calling accurate at the high polony density of the new AIVITI instrument.
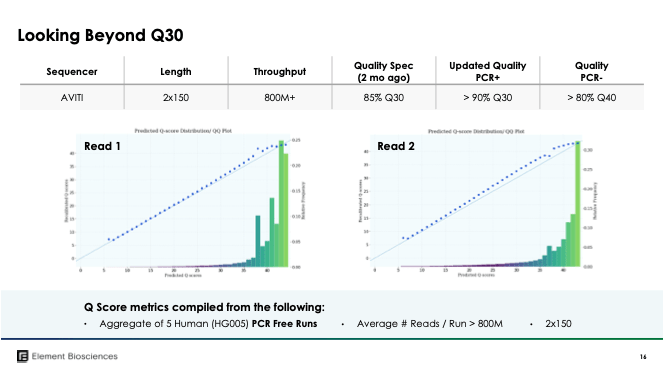
Specifications: Element are competing against Illumina’s NextSeq on multiple fronts with the AVITI instrument costing $289k versus $335k for NextSeq, delivering $5-7 per Gb run costs, compared to $16.5 on NextSeq and generating 90% Q30 at 2×150 and 80% Q40 for PCR-free libraries compared to 75% Q30 at 2 × 150 bp on NextSeq. NextSeq does have a higher output per flowcell but you can run two flowcells on AVITI and at lower overall cost to get more data in less time- this should all excite new users and Illumina NGS labs. See table below for a brief comparison of AVITI to other sequencers.
Two flowcells can be run and are randomly accessible with 800M reads each – flowcells are much more like NovaSeq in having almost hot-swappable flexibility. Additionally, the user can select how much of the flowcell to image to reduce run times for those occasions when data is needed fast – a 2x150bp 1/8th scan takes just 31 hours compared to the 48h standard run and still generates 100M reads per flowcell. This gives users the option of MiSeq-like speed and performance, albeit as higher $/Gb as reagent kit will be fully utilised even if only a portion of the flowcell is being imaged.

Modified from Albert Villela at http://bit.ly/ngsspecs
Applications and data overview
It sounds very much like there are unlikely to be any surprises for Illumina library-prep users and there some opportunity for new library-prep development due to polony amplification pre-sequencing not being bridge amplification based.
Multiple datasets are available as application notes for download from the Element Biosciences website including: WGS, WES, targeted sequencing panels, bulk RNA-Seq, single-cell RNA-Seq, etc. And 5 data sets are downloadable including multiple human whole genomes and RNA-seq (if you are reviewing data quality please get in touch if you’d like to write a guest post?). NB: I am disappointed that so little data is available at launch, releasing a 100 genomes dataset should have been possible and could have been targeted to the applications that users are doing most of.
Whilst Element have been developing their own ELEVATE library-prep chemistry they have also been busy signing up partners for the various library prep applications and downstream bioinformatics analysis. At launch the Element sequencer will be compatible with: Roche’s Kapa, Agilent Technologies, New England Biolabs, Qiagen, Watchmaker Genomics, Jumpcode Genomics, Loop Genomics (see below) and 10X Genomics. And new analysis partners include Sentieon, Google (DeepVariant), Fabric Genomics (clinical interpretation) and Genoox (Rare disease interpretation).
10X Genomics will be data many users will be interested to see. Everyone sees the power of single-cell but most of us are limited by sequencing budgets to smaller experiments than we’d like to do. As such, data from 10X, and ideally the full suite of Chromium and Visium applications (3′, 5′, TCR/BCR, spatial, etc) is probably the most impactful for signing up new customers! Check out the whitepaper/appnote on AVITI:10X compatability and there should be something similar coming soon for spatial genomics applications.
Like Illumina’s acquisition of MorphoSeq, Element Biosciences announced their acquisition of a synthetic long-read company, Loop Genomics, last month. The aim is similar, to integrate Loop’s long-read technology with Element’s short-read sequencer and make both short- and long-reads available to their users – presumably at lower cost and hopefully even easier than Ilumina can deliver? Personally, I’m yet to be convinced that people wanting long-reads shouldn’t just go and use a long-read sequencer!
Illumina library conversion: Understanding how a user can convert an Illumina library to be Element-compatible is going to be important for many early adopters. Element said that most ILMN libraries can be converted using their ADEPT conversion chemistry (a few more details are included in their app note) but all dsDNA libraries but not some single-strand libraries. What we do know is that ADEPT conversion is not an amplification but rather a simple 75 min, 15 min hands-on, protocol including a cleanup. If conversion is this easy then people with big 10X libraries, exomes, RNA-Seq etc will be able to get a very quick idea of how well Element can deliver their sequencing needs. And they’ll have confidence that if they need to stick with a home-brew Illumina method (there are over 400 out there) and convert, that this won’t cause too many issues in their downstream analysis and interpretation e.g. increased variant calling background. Incidentally, index-hopping is not a problem based on their analysis of internal data.
The library prep methods described at launch have plenty of detail, which should enable users to understand how easy (or difficult) it might be to switch from Illumina-based methods.
Sequencing data availability: The data presented in the Element Biosciences lunch today includes multiple human genomes (see figure below). This includes data from the very well sequenced GIAB samples e.g. NA12878/HG001, with over 1.6B reads generated in each run with >90% Q30 reads (highlighting that quality of Element Biosciences is unlikely to be an issue for new users). RNA-Seq data were highly concordant between ILMN and ELEM R=.995 at SE50 and PE75. Metagenomics data were similaryl concordant (R=.9998 and abundance analysis was very clearly concordant).
Downstream analysis appears to use standard tools like GATK, VARDICT, MUTECT, etc all of which accept Element data without too much effort in modifying your bioinformatics workflows and FastQ is the same file format as ILMN. All of this means that inputs to informatics pipelines should be very smooth. Similar to library-prep methods (see above), bioinformatics methods appear to be using pretty standard tools, which should mean very few changes to standard pipelines.
Early access labs will have data available at launch. I expect to get more detail from the Element launch presentation as some of their early-access users are likely to reveal additional details – so watch for an update.
The competitive landscape for short-read sequencers
As mentioned in the introduction Illumina has multiple competitors in both the short- and (debatably) long-read space e.g. BGI’s MGI, Singular Genomics, Genapsys, PacBio’s Omniome, ONT’s short-fragment-mode (SFM), et al. And this means Element Biosciences has similarly sized competition so it’s not just Illumina they’ve got to be concerned about.
The additional flexibility of their AVIDITY sequencing chemistry might prove to be important in the long run. But Illumina’s new Chemistry X (still SBS as far as we know) might yet throw a spanner in the works; possibly developed after the Element team left Illumina and, as it’s new, Element et al can’t easily fast-follow if the IP stops them.
Whether the Element Biosciences AVITI technology is clearly enough differentiated from Illumina, AND has a strong enough IP position to survive (presumed) Illumina lititgation, AND has enough of a unique value proposition for their technology and user applications remains to be seen. Ultimately, Element Biosciences need to convince users to switch from Illumina (which may be hard) or not buy something else e.g. BGI (who are geopolitically challenged), Singular (who are probably not differentiated enough from Illumina) or one of the long-read-short-read options like PacBio’s Omniome or ONTs SFR (I’m still not really clear about how these will take market share from other short-read providers) to drive adoption of their platform and provide the revenue growth and margin expansion their investors will be looking for.
Final summary
Element Bioscience, similar to Singular Genomics, are entering a $5 billion NGS market that has been starved of competition for almost a decade. Today this market is owned by Illumina (>95% of all NGS generated and at least >75% market share). Whether the GRAIL acquisition has done enough damage to Illumina’s relationships with other cancer genomics vendors e.g. Foundation Medicine, Guardant Health, Invitae to make them switch to Element Bioscience or not remains to be seen. But, and again somewhat similar to Singular Genomics, the Element AVITI technology does not deliver the high-throughput NovaSeq challenger these companies might want…yet!
Nobody got fired for buying Illumina
Wise old sage
A major question is how many of the smaller Illumina users will switch from NextSeq to AVITI? Element are aiming to sell to anyone using or considering Illumina’s desktop NextSeq instrument. This suggests they’ll be targeting individual PIs and core labs aiming to sell them a better/faster/cheaper machine at a price/throughput around the low end of SP or S1 on NovaSeq (but which costs three times as much in CapEx). The thinking process of these users will be driven by how easy the switch is and how much money will it cost/save. The financials so far disclosed look pretty attractive so I expect a reasonable number of users will migrate. This is quite different to what Singular genomics have pitched (“same price but more flexibility”) where I’m less certain there’s enough value for users compared to the risk. However, “Nobody got fired for buying Illumina” remains a challenge Elements marketing team need to respond to. If the applications space is well covered this will help core-labs who need to offer a wide variety of methods on a single instrument. Again, if the library-prep (or conversion) and informatics methods are highly Illumina-compatible then this will be a big plus-point for people thinking about switching over.
Expect to hear from more of the early access users post-launch and look out for those first Element Biosciences publications.
The next 12 months will be interesting to say the least. Will Element Biosciences place over 100 instruments before the end of the year? Who else will make announcements at, or before, the delayed AGBT? Will Illumina’s Chemistry X and Morphoseq play in long-reads be enough to beat off this competition?








Hey James,
Might want to check your yield specs for P3 and SP flow cells. Also NextSeq 1K and 2K specs are 85% >Q30 these days.
Cheers
Excellent content! Enjoy all the articles, well done!
Thanks Barbara. Pleased to make your acquaintance.