Two papers in Science last week show how CRISPR can be used as a diagnostic tool. The first comes from the same team at The Broad that brought us the SHERLOCK method, the second from Jennifer Dounda’s group at Berkley. Both methods promise simple diagnostics from technology developed for genome engineering.
The Broad Institute:
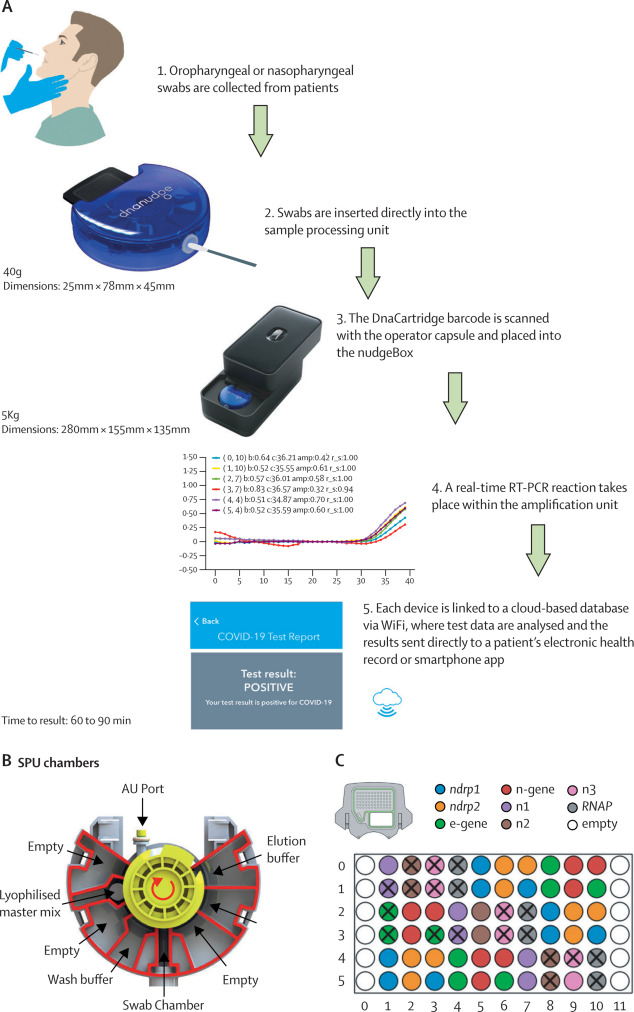
In Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6 (Science Feb 2018): Feng Zhang’s group report on their updated SHERLOCK method, craftily named SHERLOCKv2. This uses a combination of CRISPR enzymes (making full use of the unique cleavage preferences of different Cas enzymes) multiplexed in four channels (LwaCas13a, PsmCas13b, CcaCas13b, and AsCas12a in FAM, TEX, Cy5, and HEX respectively). Detection of targets was possible down to 2aM, although the paper does report the requirement for an amplification (RPA) to allow single molecule detection of all targets. The multiplex test uses a visual readout on lateral flow strips (similar to a pregnancy test): CRIPSR destruction of a FAM-biotin reporter in the presence of target molecules means the simple positive/negative result can be readout in just 90 minutes. They tested for ZIKV or DENV ssRNA detection at 2 aM concentration. They sowed thatrapid genotyping could be achieved after a 10 min DNA extraction from saliva with no additional purification in under 23 min (fluorescence) or 2 hours (lateral flow).

In CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity (Science Feb 2018): Jennifer Dounda’s group report on an almost identical method DETECTR (DNA Endonuclease Targeted CRISPR Trans Reporter), which uses Cas12a ssDNase with isothermal amplification to deliver attomolar sensitivity for rapid and specific detection of HPV.
They report results from DNA extracted from cultured human cells infected with HPV types 16 (SiHa), 18 (HeLa), or without HPV (BJAB). DETECTR unambiguously identified HPV types 16 and 18 only in SiHa and HeLa cells, respectively. They followed up this with tests on crude DNA extractions from 25 human anal swabs previously analysed by a PCR. DETECTR accurately identified HPV16 (25/25 agreement) and HPV18 (23/25 agreement) in patient samples containing a heterogeneous mixture of HPV types, with good correlation to PCR.
Summary:
These results from two groups working on very similar methods show that a new molecular diagnostic may be ready to start testing in the clinic. There’s sure to be lots more to work out on the methodologies but a simple workflow and simple readout from a test that conceivably could be made for pennies (or at least >$10) is likely to spur lots more development in this area. Time will tell if this is another CRISPR success or an interesting, but ultimately unusable, implementation of this technology.
Time will also tell if another patent battle is going to start between these two groups!






CRISPR: DETECTR The first study comes from Jennifer Doudna, an American biochemist and leading figure of the CRISPR revolution. Her team of researchers have produced a variant of the CRISPR tool that is able not only to cut through specific bits of double-stranded DNA but also snip single-stranded DNA near it. After discovering this, the team used CRISPR to detect a couple of common types of HPV. It’s called DETECTR, takes about an hour to produce results, and costs less than a dollar.
[…] With its potential already demonstrated in research, the next big milestone for CRISPR will be demonstrating to be safe and effective as a treatment. But there are many other cool applications underway. One of them is the use of CRISPR to modify the pig genome so that their organs can be transplanted into humans without creating rejection, thus circumventing donor shortage. eGenesis, co-founded by George Church, is already working on it. Another is the use of CRISPR as a diagnostics tool. […]
[…] been following CRISPR as a diagnostic tool since the first papers from Doudna/Zhang groups. CRISPR technology has been around a little […]