PAL-Seq
Poly(A)-Tail Length Profiling by Sequencing
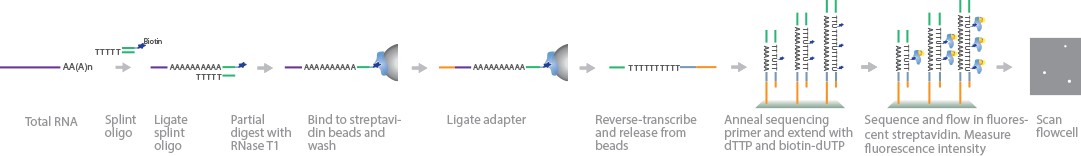
PAL-Seq measures poly(A)-tail length by incorporating fluorescent tags on biotinylated deoxyuridine triphosphate (dUTPs) and using signal intensity to quantify poly(A)-tail length (Subtelny et al., 2014). Similar to 3P-Seq RNA library preparation, a splint oligonucleotide containing a 3′-adapter sequence is ligated to the 3′ end of polyadenylated RNA and partially digested with RNase T1. To separate mRNA from total RNA, the sample is size-selected by gel purification and bound to streptavidin beads before phosphorylating the 5′ ends for adapter ligation. Before cluster generation, the mRNA fragments are reverse-transcribed into cDNA, released from the beads, and purified by size selection through a gel. Sequencing primers are attached to the 3′ end of the poly(A) sequence and extended using deoxythymidine triphosphate (dTTP) and biotinylated dUTP. To map the fragments, regions near the 5′ end of the poly(A) tails are sequenced. Fluorescent-labeled streptavidin molecules are attached to the biotin-dUTPs, and their signal intensity is measured to quantify the length of the adenine homopolymers in each cluster.
Advantages:
- Accurate measurement of poly(A)-tail length, regardless of length
- Avoids direct sequencing of the poly(A) tail
Disadvantages:
- Technically complex to perform
- Efficiency-related issues may arise during the biotin-dUTP extension step
- Only captures 3′ ends that consist purely of adenines
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
Bangru S. and Kalsotra A. Advances in analyzing RNA diversity in eukaryotic transcriptomes: peering through the Omics lens. F1000Research. 2016;5:2668
Nussbacher J. K., Batra R., Lagier-Tourenne C. and Yeo G. W. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends Neurosci. 2015;38:226-236
References:
Eichhorn S. W., Subtelny A. O., Kronja I., Kwasnieski J. C., Orr-Weaver T. L. and Bartel D. P. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. Elife. 2016;5:
Harrison P. F., Powell D. R., Clancy J. L., et al. PAT-seq: a method to study the integration of 3′-UTR dynamics with gene expression in the eukaryotic transcriptome. RNA. 2015;21:1502-1510
Kappel C., Trost G., Czesnick H., et al. Genome-Wide Analysis of PAPS1-Dependent Polyadenylation Identifies Novel Roles for Functionally Specialized Poly(A) Polymerases in Arabidopsis thaliana. PLoS Genet. 2015;11:e1005474
Related
History: PAL-Seq
Revision by sbrumpton on 2017-06-21 07:50:24 - Show/Hide
Poly(A)-Tail Length Profiling by Sequencing

PAL-Seq measures poly(A)-tail length by incorporating fluorescent tags on biotinylated deoxyuridine triphosphate (dUTPs) and using signal intensity to quantify poly(A)-tail length (Subtelny et al., 2014). Similar to 3P-Seq RNA library preparation, a splint oligonucleotide containing a 3'-adapter sequence is ligated to the 3' end of polyadenylated RNA and partially digested with RNase T1. To separate mRNA from total RNA, the sample is size-selected by gel purification and bound to streptavidin beads before phosphorylating the 5' ends for adapter ligation. Before cluster generation, the mRNA fragments are reverse-transcribed into cDNA, released from the beads, and purified by size selection through a gel. Sequencing primers are attached to the 3' end of the poly(A) sequence and extended using deoxythymidine triphosphate (dTTP) and biotinylated dUTP. To map the fragments, regions near the 5' end of the poly(A) tails are sequenced. Fluorescent-labeled streptavidin molecules are attached to the biotin-dUTPs, and their signal intensity is measured to quantify the length of the adenine homopolymers in each cluster.
Advantages:- Accurate measurement of poly(A)-tail length, regardless of length
- Avoids direct sequencing of the poly(A) tail
Disadvantages:- Technically complex to perform
- Efficiency-related issues may arise during the biotin-dUTP extension step
- Only captures 3' ends that consist purely of adenines
Reagents:Illumina Library prep and Array Kit SelectorReviews:Bangru S. and Kalsotra A. Advances in analyzing RNA diversity in eukaryotic transcriptomes: peering through the Omics lens. F1000Research. 2016;5:2668Nussbacher J. K., Batra R., Lagier-Tourenne C. and Yeo G. W. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends Neurosci. 2015;38:226-236References:Eichhorn S. W., Subtelny A. O., Kronja I., Kwasnieski J. C., Orr-Weaver T. L. and Bartel D. P. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. Elife. 2016;5:Harrison P. F., Powell D. R., Clancy J. L., et al. PAT-seq: a method to study the integration of 3'-UTR dynamics with gene expression in the eukaryotic transcriptome. RNA. 2015;21:1502-1510Kappel C., Trost G., Czesnick H., et al. Genome-Wide Analysis of PAPS1-Dependent Polyadenylation Identifies Novel Roles for Functionally Specialized Poly(A) Polymerases in Arabidopsis thaliana. PLoS Genet. 2015;11:e1005474
