CIRS-Seq
Chemical Inference of RNA Structures
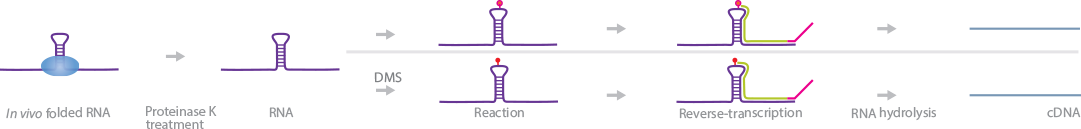
CIRS-Seq was developed to investigate the complexity of secondary RNA structures in the mammalian transcriptome (Incarnato et al., 2014). CIRS-Seq uses DMS to methylate the N1 of adenosine and N3 of cytosine residues, and CMC to modify pseudouridines selectively, but only when they are in single-stranded conformation. Modifications on these nucleotide residues halt the RT process, effectively producing a truncated cDNA as a marker for the location of secondary RNA structures.
Briefly, cells are lysed and treated with proteinase K to dissociate protein-bound RNAs while leaving RNA secondary structures intact. The lysates are separated into 3 different treatment linesãDMS, CMC, and no treatment. In all 3 treatment lines, total RNA is extracted and reverse-transcribed using random primers. The resulting cDNA is isolated, ligated to sequencing adapters, and subjected to high-throughput sequencing. Reads from the DMS and CMC lines are used to identify the locations of secondary RNA structures, while the control is used to reduce background noise.
Advantages:
- Accurately predicts secondary RNA structures, and reveals features of mRNAs and ncRNAs
- Provides single-base resolution
- Can identify structural requirements for RBPs
Disadvantages:
- CMC and DMS may react with non_secondary RNA structures
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
Zucchelli S., Patrucco L., Persichetti F., Gustincich S. and Cotella D. Engineering Translation in Mammalian Cell Factories to Increase Protein Yield: The Unexpected Use of Long Non-Coding SINEUP RNAs. Comput Struct Biotechnol J. 2016;14:404-410
Kwok C. K., Tang Y., Assmann S. M. and Bevilacqua P. C. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem Sci. 2015;40:221-232
Strobel E. J., Watters K. E., Loughrey D. and Lucks J. B. RNA systems biology: uniting functional discoveries and structural tools to understand global roles of RNAs. Curr Opin Biotechnol. 2016;39:182-191
References:
Incarnato D., Anselmi F., Morandi E., et al. High-throughput single-base resolution mapping of RNA 2′-O-methylated residues. Nucleic Acids Res. 2016;
Related
History: CIRS-Seq
Revision by sbrumpton on 2017-06-21 09:06:28 - Show/Hide
Chemical Inference of RNA Structures

CIRS-Seq was developed to investigate the complexity of secondary RNA structures in the mammalian transcriptome (Incarnato et al., 2014). CIRS-Seq uses DMS to methylate the N1 of adenosine and N3 of cytosine residues, and CMC to modify pseudouridines selectively, but only when they are in single-stranded conformation. Modifications on these nucleotide residues halt the RT process, effectively producing a truncated cDNA as a marker for the location of secondary RNA structures.
Briefly, cells are lysed and treated with proteinase K to dissociate protein-bound RNAs while leaving RNA secondary structures intact. The lysates are separated into 3 different treatment linesãDMS, CMC, and no treatment. In all 3 treatment lines, total RNA is extracted and reverse-transcribed using random primers. The resulting cDNA is isolated, ligated to sequencing adapters, and subjected to high-throughput sequencing. Reads from the DMS and CMC lines are used to identify the locations of secondary RNA structures, while the control is used to reduce background noise.
Advantages:- Accurately predicts secondary RNA structures, and reveals features of mRNAs and ncRNAs
- Provides single-base resolution
- Can identify structural requirements for RBPs
Disadvantages:- CMC and DMS may react with non_secondary RNA structures
Reagents:Illumina Library prep and Array Kit SelectorReviews:Zucchelli S., Patrucco L., Persichetti F., Gustincich S. and Cotella D. Engineering Translation in Mammalian Cell Factories to Increase Protein Yield: The Unexpected Use of Long Non-Coding SINEUP RNAs. Comput Struct Biotechnol J. 2016;14:404-410 Kwok C. K., Tang Y., Assmann S. M. and Bevilacqua P. C. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem Sci. 2015;40:221-232Strobel E. J., Watters K. E., Loughrey D. and Lucks J. B. RNA systems biology: uniting functional discoveries and structural tools to understand global roles of RNAs. Curr Opin Biotechnol. 2016;39:182-191References:Incarnato D., Anselmi F., Morandi E., et al. High-throughput single-base resolution mapping of RNA 2'-O-methylated residues. Nucleic Acids Res. 2016;
