HiTS-RAP
High-Throughput SequencingãRNA Affinity Profiling
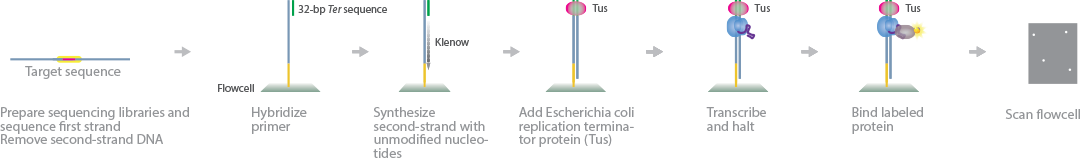
HiTS-RAP is a quantitative method to evaluate binding interactions of RNA aptamers with their proteins at a massive scale (Tome et al., 2014) (Ozer et al., 2015). HiTS-RAP transcribes DNA directly on the flow cell and measures the binding affinity of RNA aptamers with fluorescently labeled proteins.
First, sequencing libraries are prepared and sequenced on the Illumina flow cell. Next, the second strand of the DNA is removed and the single-stranded DNA attached to the flow cell is annealed to primers containing a 32 bp Ter sequence. An Escherichia coli replication terminator protein (Tus) is introduced into the system and binds to the Ter sequence. RNA transcription by T7 RNA polymerase begins and halts upstream of the Tus-bound Ter site. Because the RNA polymerase activity was halted abruptly due to the Tus bound to the DNA, the RNA strand is still linked to the RNA polymerase complex. Fluorescently labeled proteins are introduced and bind to the exposed RNA aptamers, producing a fluorescent signal that can be read by the sequencer.
Similar methods: RBNS, RNA-MaP
Advantages:
- Quantitatively evaluates binding affinity of RNA aptamers on a massively parallel scale.
- Can be adapted easily to nonprotein molecules
- Able to determine de novo binding specificity of RBPs
Disadvantages:
- Limited to analysis of RNA fragments 150 nt or shorter, unless paired-end sequencing protocols are used, which increases the limit to 500 nt
- Unable to measure binding kinetics, such as on and off rates
- Susceptible to steric hindrance effects usually present in large proteins
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
Marchese D., de Groot N. S., Lorenzo Gotor N., Livi C. M. and Tartaglia G. G. Advances in the characterization of RNA-binding proteins. Wiley Interdiscip Rev RNA. 2016;
Campbell Z. T. and Wickens M. Probing RNA-protein networks: biochemistry meets genomics. Trends Biochem Sci. 2015;40:157-164
References:
Tome J. M., Ozer A., Pagano J. M., Gheba D., Schroth G. P. and Lis J. T. Comprehensive analysis of RNA-protein interactions by high-throughput sequencing-RNA affinity profiling. Nat Methods. 2014;11:683-688
Related
History: HiTS-RAP
Revision by sbrumpton on 2017-06-21 09:06:28 - Show/Hide
High-Throughput SequencingãRNA Affinity Profiling

HiTS-RAP is a quantitative method to evaluate binding interactions of RNA aptamers with their proteins at a massive scale (Tome et al., 2014) (Ozer et al., 2015). HiTS-RAP transcribes DNA directly on the flow cell and measures the binding affinity of RNA aptamers with fluorescently labeled proteins.
First, sequencing libraries are prepared and sequenced on the Illumina flow cell. Next, the second strand of the DNA is removed and the single-stranded DNA attached to the flow cell is annealed to primers containing a 32 bp Ter sequence. An Escherichia coli replication terminator protein (Tus) is introduced into the system and binds to the Ter sequence. RNA transcription by T7 RNA polymerase begins and halts upstream of the Tus-bound Ter site. Because the RNA polymerase activity was halted abruptly due to the Tus bound to the DNA, the RNA strand is still linked to the RNA polymerase complex. Fluorescently labeled proteins are introduced and bind to the exposed RNA aptamers, producing a fluorescent signal that can be read by the sequencer.
Similar methods: RBNS, RNA-MaP
Advantages:- Quantitatively evaluates binding affinity of RNA aptamers on a massively parallel scale.
- Can be adapted easily to nonprotein molecules
- Able to determine de novo binding specificity of RBPs
Disadvantages:- Limited to analysis of RNA fragments 150 nt or shorter, unless paired-end sequencing protocols are used, which increases the limit to 500 nt
- Unable to measure binding kinetics, such as on and off rates
- Susceptible to steric hindrance effects usually present in large proteins
Reagents:Illumina Library prep and Array Kit SelectorReviews:Marchese D., de Groot N. S., Lorenzo Gotor N., Livi C. M. and Tartaglia G. G. Advances in the characterization of RNA-binding proteins. Wiley Interdiscip Rev RNA. 2016;Campbell Z. T. and Wickens M. Probing RNA-protein networks: biochemistry meets genomics. Trends Biochem Sci. 2015;40:157-164References:Tome J. M., Ozer A., Pagano J. M., Gheba D., Schroth G. P. and Lis J. T. Comprehensive analysis of RNA-protein interactions by high-throughput sequencing-RNA affinity profiling. Nat Methods. 2014;11:683-688
