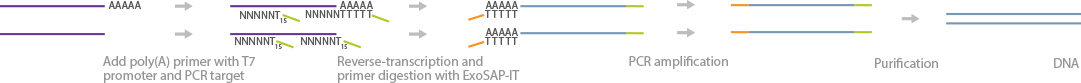SUPeR-Seq
Single-Cell Universal Poly(A)-Independent RNA Sequencing
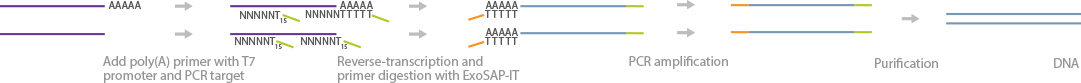
SUPeR-Seq sequences non-poly(A) and poly(A) RNAs from single cells. It is designed particularly for mapping circular RNA (circRNA) species (Fan et al., 2015). RNA samples from lysed single cells are annealed to random primers with universal anchor sequences (AnchorX-T15N6) and reverse-transcribed to generate the first strand of cDNA. Unreacted primers are digested, to avoid primer-dimers. Next, a poly(A) tract is added to the 3′ end of the cDNA by introducing dATP and ddATP in a 100:1 ratio, respectively. A second set of random primers, also with a universal anchor sequence (AnchorY-T24), anneals to the newly synthesized poly(A) tract. A second cDNA strand is generated by RT, and the cDNA is purified by gel electrophoresis. The purified cDNA molecules are PCR-amplified using 5′-amine-terminated primers, prepared by the TruSeq DNA library preparation protocol, and sequenced. circRNAs are identified from the dataset by finding 2 exonic reads that are distal in the reference genome, but adjacent to each other in the dataset with 1 inverted, signifying the circularization of the RNA.
Advantages:
- Identifies circRNAs from single cells
- Avoids 3′ bias by using random primers with anchor sequences
- Able to identify novel circRNAs due to random primers
Disadvantages:
- Relies on dataset analysis to identify circRNAs
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
No reviews yet
References:
Dang Y., Yan L., Hu B., et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016;17:130
Related
History: SUPeR-Seq
Revision by sbrumpton on 2017-06-21 07:50:22 - Show/Hide
Single-Cell Universal Poly(A)-Independent RNA Sequencing
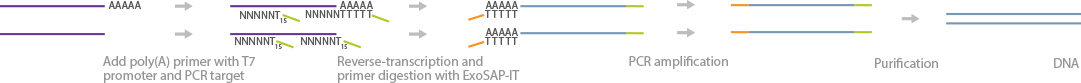
SUPeR-Seq sequences non-poly(A) and poly(A) RNAs from single cells. It is designed particularly for mapping circular RNA (circRNA) species (Fan et al., 2015). RNA samples from lysed single cells are annealed to random primers with universal anchor sequences (AnchorX-T15N6) and reverse-transcribed to generate the first strand of cDNA. Unreacted primers are digested, to avoid primer-dimers. Next, a poly(A) tract is added to the 3' end of the cDNA by introducing dATP and ddATP in a 100:1 ratio, respectively. A second set of random primers, also with a universal anchor sequence (AnchorY-T24), anneals to the newly synthesized poly(A) tract. A second cDNA strand is generated by RT, and the cDNA is purified by gel electrophoresis. The purified cDNA molecules are PCR-amplified using 5'-amine-terminated primers, prepared by the TruSeq DNA library preparation protocol, and sequenced. circRNAs are identified from the dataset by finding 2 exonic reads that are distal in the reference genome, but adjacent to each other in the dataset with 1 inverted, signifying the circularization of the RNA.
Advantages:- Identifies circRNAs from single cells
- Avoids 3' bias by using random primers with anchor sequences
- Able to identify novel circRNAs due to random primers
Disadvantages:- Relies on dataset analysis to identify circRNAs
Reagents:Illumina Library prep and Array Kit SelectorReviews:No reviews yet
References:Dang Y., Yan L., Hu B., et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016;17:130