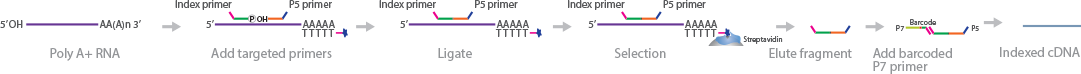RASL-Seq
RNA-Mediated Oligonucleotide Annealing, Selection, and Ligation with Next-Generation Sequencing
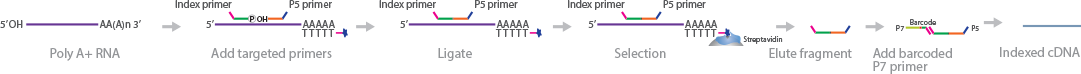
RNA-mediated oligonucleotide annealing, selection, and ligation with next-generation sequencing (RASL-Seq) is a 2-dimensional RNA sequencing method to quantify expression profiles of several hundred genes, under thousands of different conditions (Li et al., 2012). Custom probe pairs are designed for each gene of interest. A pair of probes needs to contain: 1) One probe containing a common index primer on its 3′ end and a 20 nt oligonucleotide corresponding to the targeted exon sequence with a phosphate on the 5′ end; and 2) another probe with a P5 adapter on its 5′ end with a 20 nt sequence complementary to the exon that is adjacent to the other probe. The probe pairs are hybridized to the mRNA and separated from total RNA using oligo(dT)-biotin beads. A ligation step joins the probe pairs into a single PCR amplicon probe. The biotinylated mRNA strands are subsequently attached to streptavidin magnetic beads to elute the probe fragments. Next, P7 adapters are attached to the 3′ index primer, and the library undergoes PCR amplification before sequencing. The library is sequenced from the 40 nt ligated P5 primer, followed by sequencing from the P7 primer oligonucleotide.
Advantages:
- Quantify genetic expression in large gene panels under thousands of different experimental conditions
- Effective on low total RNA amounts (10 ng for about 1000 cells)
- Can be performed on isolated RNA samples or cell lysates
- Can be performed manually or automatically using a custom robot
Disadvantages:
- Sequencing is carried out on short 40 nt fragments, which can cause problems in repetitive regions
- High rate of random ligations can occur at low RNA levels
- Automated process produces less robust results, due to random ligations during probe removal
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
Nussbacher J. K., Batra R., Lagier-Tourenne C. and Yeo G. W. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends Neurosci. 2015;38:226-236
References:
Qiu J., Zhou B., Thol F., et al. Distinct splicing signatures affect converged pathways in myelodysplastic syndrome patients carrying mutations in different splicing regulators. RNA. 2016;
Kralovicova J., Knut M., Cross N. C. and Vorechovsky I. Identification of U2AF(35)-dependent exons by RNA-Seq reveals a link between 3′ splice-site organization and activity of U2AF-related proteins. Nucleic Acids Res. 2015;
Shao C., Yang B., Wu T., et al. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat Struct Mol Biol. 2014;21:997-1005
Zhou Z., Qiu J., Liu W., et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell. 2012;47:422-433
Related
History: RASL-Seq
Revision by sbrumpton on 2017-06-21 07:50:24 - Show/Hide
RNA-Mediated Oligonucleotide Annealing, Selection, and Ligation with Next-Generation Sequencing
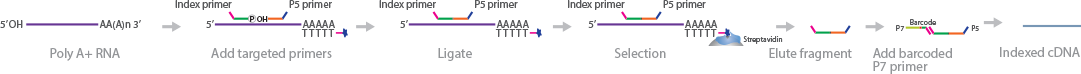
RNA-mediated oligonucleotide annealing, selection, and ligation with next-generation sequencing (RASL-Seq) is a 2-dimensional RNA sequencing method to quantify expression profiles of several hundred genes, under thousands of different conditions (Li et al., 2012). Custom probe pairs are designed for each gene of interest. A pair of probes needs to contain: 1) One probe containing a common index primer on its 3' end and a 20 nt oligonucleotide corresponding to the targeted exon sequence with a phosphate on the 5' end; and 2) another probe with a P5 adapter on its 5' end with a 20 nt sequence complementary to the exon that is adjacent to the other probe. The probe pairs are hybridized to the mRNA and separated from total RNA using oligo(dT)-biotin beads. A ligation step joins the probe pairs into a single PCR amplicon probe. The biotinylated mRNA strands are subsequently attached to streptavidin magnetic beads to elute the probe fragments. Next, P7 adapters are attached to the 3' index primer, and the library undergoes PCR amplification before sequencing. The library is sequenced from the 40 nt ligated P5 primer, followed by sequencing from the P7 primer oligonucleotide.
Advantages:- Quantify genetic expression in large gene panels under thousands of different experimental conditions
- Effective on low total RNA amounts (10 ng for about 1000 cells)
- Can be performed on isolated RNA samples or cell lysates
- Can be performed manually or automatically using a custom robot
Disadvantages:- Sequencing is carried out on short 40 nt fragments, which can cause problems in repetitive regions
- High rate of random ligations can occur at low RNA levels
- Automated process produces less robust results, due to random ligations during probe removal
Reagents:Illumina Library prep and Array Kit SelectorReviews:Nussbacher J. K., Batra R., Lagier-Tourenne C. and Yeo G. W. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends Neurosci. 2015;38:226-236References:Qiu J., Zhou B., Thol F., et al. Distinct splicing signatures affect converged pathways in myelodysplastic syndrome patients carrying mutations in different splicing regulators. RNA. 2016;Kralovicova J., Knut M., Cross N. C. and Vorechovsky I. Identification of U2AF(35)-dependent exons by RNA-Seq reveals a link between 3' splice-site organization and activity of U2AF-related proteins. Nucleic Acids Res. 2015;Shao C., Yang B., Wu T., et al. Mechanisms for U2AF to define 3' splice sites and regulate alternative splicing in the human genome. Nat Struct Mol Biol. 2014;21:997-1005Zhou Z., Qiu J., Liu W., et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell. 2012;47:422-433