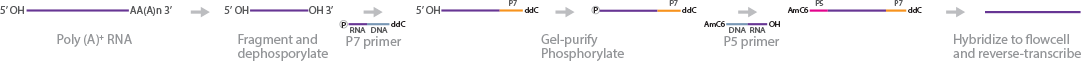FRT-Seq
Flowcell Reverse Transcription Sequencing
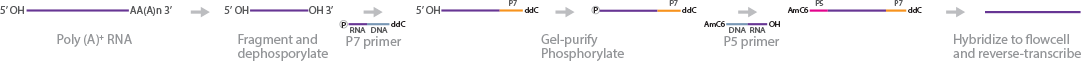
Flow-cell surface reverse transcription sequencing (FRT-Seq) is an transcriptome sequencing technique developed in 2010 by Mamanova et al (Mamanova et al., 2010). The method is strand-specific, free of amplification, and compatible with paired-end sequencing. To begin with, poly(A) RNA samples are fragmented by metal-ion hydrolysis and dephosphorylated. Next, P7 primer/adapters are ligated to the 3′ end of the fragments. The 3′ adapter sequence starts at the 5′ terminus with 20 nt of RNA followed by DNA nucleotides. The adapters are also 5′ phosphorylated and blocked with dideoxycytosine (ddC) at the 3′ end. After 3′ adapter ligation, the fragments are size-selected for nucleotide fragments longer than the adapter. The 5′ ends of the fragments are phosphorylated and ligated to P5 adapters. These adapters are blocked with an amino-C6 linker at the 5′ end. Now that the fragments are flanked by adapters, they are hybridized to the flow cell and reverse-transcribed before cluster generation and sequencing.
Advantages:
- Strand-specific poly(A) mRNA sequencing for transcriptome analysis
- No amplification stepãgives more accurate representation of the total mRNA population, preventing amplification bias
Disadvantages:
- Requires a large amount of poly(A) RNA material (250 ng)
- Selects only poly(A) mRNA samples
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
van Dijk E. L., Jaszczyszyn Y. and Thermes C. Library preparation methods for next-generation sequencing: tone down the bias. Exp Cell Res. 2014;322:12-20
McGettigan P. A. Transcriptomics in the RNA-seq era. Curr Opin Chem Biol. 2013;17:4-11
References:
Vergara-Irigaray M., Fookes M. C., Thomson N. R. and Tang C. M. RNA-seq analysis of the influence of anaerobiosis and FNR on Shigella flexneri. BMC Genomics. 2014;15:438
Mills J. D., Kawahara Y. and Janitz M. Strand-Specific RNA-Seq Provides Greater Resolution of Transcriptome Profiling. Curr Genomics. 2013;14:173-181
Kroger C., Dillon S. C., Cameron A. D., et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A. 2012;109:E1277-1286
Related
History: FRT-Seq
Revision by sbrumpton on 2017-06-21 09:06:28 - Show/Hide
Flowcell Reverse Transcription Sequencing
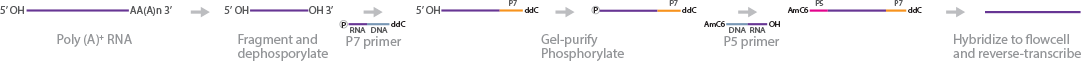
Flow-cell surface reverse transcription sequencing (FRT-Seq) is an transcriptome sequencing technique developed in 2010 by Mamanova et al (Mamanova et al., 2010). The method is strand-specific, free of amplification, and compatible with paired-end sequencing. To begin with, poly(A) RNA samples are fragmented by metal-ion hydrolysis and dephosphorylated. Next, P7 primer/adapters are ligated to the 3' end of the fragments. The 3' adapter sequence starts at the 5' terminus with 20 nt of RNA followed by DNA nucleotides. The adapters are also 5' phosphorylated and blocked with dideoxycytosine (ddC) at the 3' end. After 3' adapter ligation, the fragments are size-selected for nucleotide fragments longer than the adapter. The 5' ends of the fragments are phosphorylated and ligated to P5 adapters. These adapters are blocked with an amino-C6 linker at the 5' end. Now that the fragments are flanked by adapters, they are hybridized to the flow cell and reverse-transcribed before cluster generation and sequencing.
Advantages:- Strand-specific poly(A) mRNA sequencing for transcriptome analysis
- No amplification stepãgives more accurate representation of the total mRNA population, preventing amplification bias
Disadvantages:- Requires a large amount of poly(A) RNA material (250 ng)
- Selects only poly(A) mRNA samples
Reagents:Illumina Library prep and Array Kit SelectorReviews:van Dijk E. L., Jaszczyszyn Y. and Thermes C. Library preparation methods for next-generation sequencing: tone down the bias. Exp Cell Res. 2014;322:12-20McGettigan P. A. Transcriptomics in the RNA-seq era. Curr Opin Chem Biol. 2013;17:4-11References:Vergara-Irigaray M., Fookes M. C., Thomson N. R. and Tang C. M. RNA-seq analysis of the influence of anaerobiosis and FNR on Shigella flexneri. BMC Genomics. 2014;15:438Mills J. D., Kawahara Y. and Janitz M. Strand-Specific RNA-Seq Provides Greater Resolution of Transcriptome Profiling. Curr Genomics. 2013;14:173-181Kroger C., Dillon S. C., Cameron A. D., et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A. 2012;109:E1277-1286