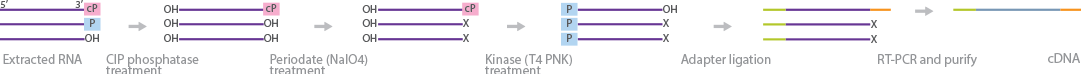cP-RNA-Seq
2′,3′-cyclic phosphate (cP) RNA Sequencing
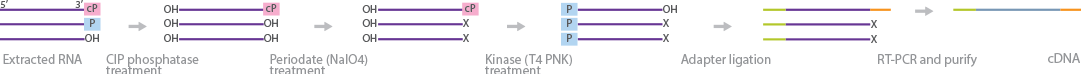
2′,3′-cyclic phosphate (cP) RNA sequencing (cP-RNA-Seq) is a method to isolate and sequence RNA species protected by cP at their 3′ terminus, which usually prevents adapter ligation (Honda et al., 2016). The researchers originally developed this technique to identify transfer RNA (tRNA) species that generate sex hormone_dependent tRNA-derived RNAs (SHOT-RNAs) in human breast cancer cells, but it can be used for other RNA species protected by cP at the 3′ end (Honda et al., 2015). Total RNA from samples is isolated and gel-purified to the desired length. This mixture contains RNA species with 3′ hydroxy, 3’phosphate, and 3′-cP. Phosphatase treatment removes phosphates from the 5′ and 3′ ends. The RNA is treated subsequently with periodate to cleave the 3′-hydroxyl ends into 2’3′-dialdehydes, while leaving the 3′-cP ends intact. RNA strands with 2’3′-dialdehydes at their 3′ terminus are inert to adapter ligation. Strands with 3′-cP are cleaved with T4 polynucleotide kinase (PNK) and ligated to sequencing adapters. After flanking both ends with adapters, the RNA strands are reverse-transcribed, amplified, purified, and sequenced.
Advantages:
- Selective isolation, amplification, and sequencing of RNA species protected by cP
- Considerable improvement in efficiency and specificity compared to an earlier method using recombinant tRNA ligase and multiple gel purifications (Honda et al., 2016).
Disadvantages:
- False positives will arise when sequencing RNA species with 2′-O-methylribose modificationsãsuch as plant miRNAs, plant and animal short interfering RNAs (siRNAs), and animal PIWI-interacting RNAs (piRNAs)ãdue to periodate cleavage specificity
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
None available yet
References:
Honda S., Loher P., Shigematsu M., et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A. 2015;112:E3816-3825
Related
History: cP-RNA-Seq
Revision by sbrumpton on 2017-06-21 09:06:28 - Show/Hide
2',3'-cyclic phosphate (cP) RNA Sequencing
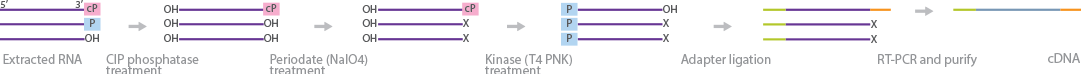
2',3'-cyclic phosphate (cP) RNA sequencing (cP-RNA-Seq) is a method to isolate and sequence RNA species protected by cP at their 3' terminus, which usually prevents adapter ligation (Honda et al., 2016). The researchers originally developed this technique to identify transfer RNA (tRNA) species that generate sex hormone_dependent tRNA-derived RNAs (SHOT-RNAs) in human breast cancer cells, but it can be used for other RNA species protected by cP at the 3' end (Honda et al., 2015). Total RNA from samples is isolated and gel-purified to the desired length. This mixture contains RNA species with 3' hydroxy, 3'phosphate, and 3'-cP. Phosphatase treatment removes phosphates from the 5' and 3' ends. The RNA is treated subsequently with periodate to cleave the 3'-hydroxyl ends into 2'3'-dialdehydes, while leaving the 3'-cP ends intact. RNA strands with 2'3'-dialdehydes at their 3' terminus are inert to adapter ligation. Strands with 3'-cP are cleaved with T4 polynucleotide kinase (PNK) and ligated to sequencing adapters. After flanking both ends with adapters, the RNA strands are reverse-transcribed, amplified, purified, and sequenced.
Advantages:- Selective isolation, amplification, and sequencing of RNA species protected by cP
- Considerable improvement in efficiency and specificity compared to an earlier method using recombinant tRNA ligase and multiple gel purifications (Honda et al., 2016).
Disadvantages:- False positives will arise when sequencing RNA species with 2'-O-methylribose modificationsãsuch as plant miRNAs, plant and animal short interfering RNAs (siRNAs), and animal PIWI-interacting RNAs (piRNAs)ãdue to periodate cleavage specificity
Reagents:Illumina Library prep and Array Kit SelectorReviews:None available yet
References:Honda S., Loher P., Shigematsu M., et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A. 2015;112:E3816-3825