RBNS
RNA Bind-n-Seq
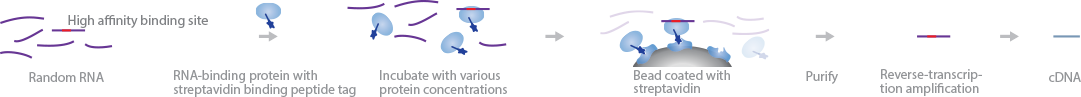
RBNS characterizes RBPs by high-throughput quantification of their binding affinity, dissociation constants, and the effects of secondary RNA structures on binding (Lambert et al., 2014). RBNS performs deep sequencing on random short RNA oligonucleotides bound to pools of fluorescently labeled RBPs of different concentrations.
First, RBPs with streptavidin tags are separated into different concentrations pools. Next, they are added to a pool of random RNA fragments. Each RNA oligonucleotide is made up of high- and low-affinity binding sites that are flanked by sequencing adapters at both ends. After the RBPs bind to their target RNAs, the complexes are purified by streptavidin pull-down and the bound RNAs are eluted out. Standard cDNA library preparation procedures are carried out to produce the sequencing library.
Similar methods: HiTS-RAP
Advantages:
- Characterizes sequence and specificity of RBPs
- Complements crosslinking-based methods
Disadvantages:
- Only uses a single round of selection, yielding shorter core RBP sites (Campbell et al., 2015)
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
Campbell Z. T. and Wickens M. Probing RNA-protein networks: biochemistry meets genomics. Trends Biochem Sci. 2015;40:157-164
References:
Taliaferro J. M., Lambert N. J., Sudmant P. H., et al. RNA Sequence Context Effects Measured In Vitro Predict In Vivo Protein Binding and Regulation. Mol Cell. 2016;64:294-306