STRT-Seq
Single-Cell Tagged Reverse Transcription Sequencing
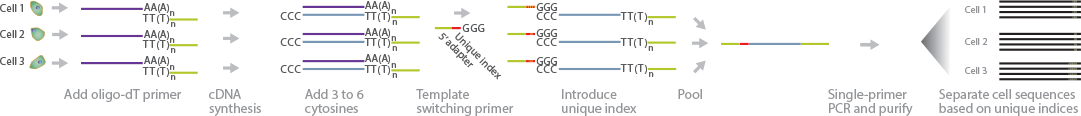
STRT-Seq is a method similar to CEL-Seq that involves unique barcoding and sample pooling to overcome the challenges of samples with limited material (Islam et al., 2011) (Islam et al., 2012). In this method, single cells are first picked in individual tubes, where first-strand cDNA synthesis occurs using an oligo(dT) primer with the addition of 3_6 cytosines. A helper oligonucleotide promotes template switching, which introduces the barcode into the cDNA. The barcoded cDNA is amplified by single-primer PCR. Deep sequencing allows for accurate transcriptome determination of individual cells.
Advantages:
- Barcoding and pooling allows for multiplexing and studying many different single cells at a time
- Sample handling and the potential for cross-contamination are greatly reduced due to using a single tube per cell
Disadvantages:
- PCR biases can underrepresent GC-rich templates
- Nonlinear PCR amplification can lead to biases affecting reproducibility
- Amplification errors caused by polymerases will be represented and sequenced incorrectly
- Loss of accuracy due to PCR bias
- Targets smaller than 500 bp are amplified preferentially by polymerases during PCR
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
Macaulay I. C. and Voet T. Single cell genomics: advances and future perspectives. PLoS Genet. 2014;10:e1004126
Kolodziejczyk A. A., Kim J. K., Svensson V., Marioni J. C. and Teichmann S. A. The Technology and Biology of Single-Cell RNA Sequencing. Mol Cell. 2015;58:610-620
Sun H. J., Chen J., Ni B., Yang X. and Wu Y. Z. Recent advances and current issues in single-cell sequencing of tumors. Cancer Lett. 2015;365:1-10
Grun D. and van Oudenaarden A. Design and Analysis of Single-Cell Sequencing Experiments. Cell. 2015;163:799-810
Zhang X., Marjani S. L., Hu Z., Weissman S. M., Pan X., et al. Single-Cell Sequencing for Precise Cancer Research: Progress and Prospects. Cancer Res. 2016;76:1305-1312
Navin N. E. Cancer genomics: one cell at a time. Genome Biol. 2014;15:452
References:
Korber I., Katayama S., Einarsdottir E., et al. Gene-Expression Profiling Suggests Impaired Signaling via the Interferon Pathway in Cstb-/- Microglia. PLoS One. 2016;11:e0158195
Tohonen V., Katayama S., Vesterlund L., et al. Novel PRD-like homeodomain transcription factors and retrotransposon elements in early human development. Nat Commun. 2015;6:8207
Katayama S., Skoog T., Jouhilahti E. M., et al. Gene expression analysis of skin grafts and cultured keratinocytes using synthetic RNA normalization reveals insights into differentiation and growth control. BMC Genomics. 2015;16:476
Related
History: STRT-Seq
Revision by sbrumpton on 2017-06-21 07:50:22 - Show/Hide
Single-Cell Tagged Reverse Transcription Sequencing

STRT-Seq is a method similar to CEL-Seq that involves unique barcoding and sample pooling to overcome the challenges of samples with limited material (Islam et al., 2011) (Islam et al., 2012). In this method, single cells are first picked in individual tubes, where first-strand cDNA synthesis occurs using an oligo(dT) primer with the addition of 3_6 cytosines. A helper oligonucleotide promotes template switching, which introduces the barcode into the cDNA. The barcoded cDNA is amplified by single-primer PCR. Deep sequencing allows for accurate transcriptome determination of individual cells.
Advantages:- Barcoding and pooling allows for multiplexing and studying many different single cells at a time
- Sample handling and the potential for cross-contamination are greatly reduced due to using a single tube per cell
Disadvantages:- PCR biases can underrepresent GC-rich templates
- Nonlinear PCR amplification can lead to biases affecting reproducibility
- Amplification errors caused by polymerases will be represented and sequenced incorrectly
- Loss of accuracy due to PCR bias
- Targets smaller than 500 bp are amplified preferentially by polymerases during PCR
Reagents:Illumina Library prep and Array Kit SelectorReviews:Macaulay I. C. and Voet T. Single cell genomics: advances and future perspectives. PLoS Genet. 2014;10:e1004126Kolodziejczyk A. A., Kim J. K., Svensson V., Marioni J. C. and Teichmann S. A. The Technology and Biology of Single-Cell RNA Sequencing. Mol Cell. 2015;58:610-620Sun H. J., Chen J., Ni B., Yang X. and Wu Y. Z. Recent advances and current issues in single-cell sequencing of tumors. Cancer Lett. 2015;365:1-10Grun D. and van Oudenaarden A. Design and Analysis of Single-Cell Sequencing Experiments. Cell. 2015;163:799-810Zhang X., Marjani S. L., Hu Z., Weissman S. M., Pan X., et al. Single-Cell Sequencing for Precise Cancer Research: Progress and Prospects. Cancer Res. 2016;76:1305-1312Navin N. E. Cancer genomics: one cell at a time. Genome Biol. 2014;15:452References:Korber I., Katayama S., Einarsdottir E., et al. Gene-Expression Profiling Suggests Impaired Signaling via the Interferon Pathway in Cstb-/- Microglia. PLoS One. 2016;11:e0158195 Tohonen V., Katayama S., Vesterlund L., et al. Novel PRD-like homeodomain transcription factors and retrotransposon elements in early human development. Nat Commun. 2015;6:8207 Katayama S., Skoog T., Jouhilahti E. M., et al. Gene expression analysis of skin grafts and cultured keratinocytes using synthetic RNA normalization reveals insights into differentiation and growth control. BMC Genomics. 2015;16:476
