OS-Seq
Oligonucleotide-Selective Sequencing
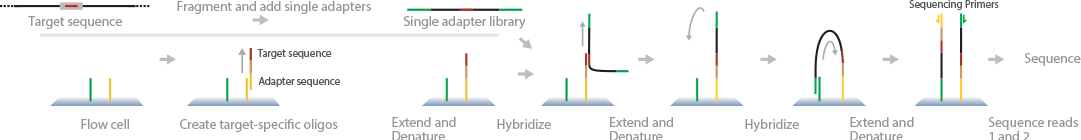
OS-Seq was developed to improve targeted resequencing by capturing and sequencing gene targets directly on the flow cell (Myllykangas et al., 2011). In this method, target sequences with adapters are used to modify the flow cell primers. Targets in the template are captured onto the flow cell with the modified primers. Further extension, denaturation, and hybridization provide sequence reads for the target genes. Deep sequencing provides accurate representation of reads.
Advantages:
- Can resequence multiple targets at a time
- No gel excision or narrow size-purification required
- Rapid, single-day protocol
- Samples can be multiplexed
- Reduced PCR bias due to removal of amplification steps
- Avoids loss of material
Disadvantages:
- Primers may interact with similar target sequences, leading to sequence ambiguity
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
None available yet
References:
Akinrinade O., Ollila L., Vattulainen S., et al. Genetics and genotype-phenotype correlations in Finnish patients with dilated cardiomyopathy. Eur Heart J. 2015;36:2327-2337
Vattulainen S., Aho J., Salmenpera P., et al. Accurate genetic diagnosis of Finnish pulmonary arterial hypertension patients using oligonucleotide-selective sequencing. Mol Genet Genomic Med. 2015;3:354-362
Hopmans E. S., Natsoulis G., Bell J. M., Grimes S. M., Sieh W. and Ji H. P. A programmable method for massively parallel targeted sequencing. Nucleic Acids Res. 2014;42:e88
Related
History: OS-Seq
Revision by sbrumpton on 2017-06-21 07:50:21 - Show/Hide
Oligonucleotide-Selective Sequencing

OS-Seq was developed to improve targeted resequencing by capturing and sequencing gene targets directly on the flow cell (Myllykangas et al., 2011). In this method, target sequences with adapters are used to modify the flow cell primers. Targets in the template are captured onto the flow cell with the modified primers. Further extension, denaturation, and hybridization provide sequence reads for the target genes. Deep sequencing provides accurate representation of reads.
Advantages:- Can resequence multiple targets at a time
- No gel excision or narrow size-purification required
- Rapid, single-day protocol
- Samples can be multiplexed
- Reduced PCR bias due to removal of amplification steps
- Avoids loss of material
Disadvantages:- Primers may interact with similar target sequences, leading to sequence ambiguity
Reagents:Illumina Library prep and Array Kit SelectorReviews:None available yet
References:Akinrinade O., Ollila L., Vattulainen S., et al. Genetics and genotype-phenotype correlations in Finnish patients with dilated cardiomyopathy. Eur Heart J. 2015;36:2327-2337Vattulainen S., Aho J., Salmenpera P., et al. Accurate genetic diagnosis of Finnish pulmonary arterial hypertension patients using oligonucleotide-selective sequencing. Mol Genet Genomic Med. 2015;3:354-362Hopmans E. S., Natsoulis G., Bell J. M., Grimes S. M., Sieh W. and Ji H. P. A programmable method for massively parallel targeted sequencing. Nucleic Acids Res. 2014;42:e88
