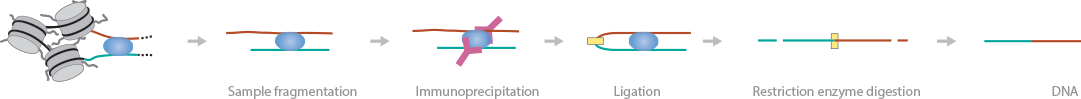ChIA-PET
Chromatin Interaction Analysis by Paired-End Tag Sequencing
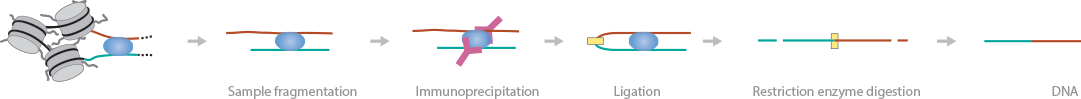
ChIA-PET features an immunoprecipitation step to map long-range DNA interactions, similar to Hi-C (Li et al., 2010) (Fullwood et al., 2009). In this method, DNA-protein complexes are crosslinked and fragmented. Specific antibodies are used to immunoprecipitate proteins of interest. Two sets of linkers, with unique barcodes, are ligated to the ends of the DNA fragments in separate aliquots, which then self-ligate based on proximity. The DNA aliquots are precipitated, digested with restriction enzymes, and sequenced. Deep sequencing provides base-pair resolution of the ligated fragments. Hi-C and ChIA-PET currently provide the best balance of resolution and reasonable coverage in the human genome to map long-range interactions (Dekker et al., 2013).
A modified protocol, called advanced or long-read ChIA-PET, has been published by Tang et al (Tang et al., 2015). This method replaces the 2 separate ligation reactions with 2 half linkers and a single biotinylated linker ligation. Next, the de-crosslinked, purified DNA is fragmented and adapters are ligated using Tn5 transposase in a single step. Finally, the DNA is PCR-amplified and sequenced (Sati et al., 2016)
Advantages:
- Suitable for detecting a large number of both long-range and short-range chromatin interactions globally (Sajan et al., 2012)
- Studies the interactions made by specific proteins or protein complexes
- Public ChIA-PET datasets are available through the ENCODE Project (Consortium et al., 2011)
- Removes background generated during traditional ChIP assays
- Immunoprecipitation step reduces data complexity
Disadvantages:
- Requires a large amount of starting material required, generally at least 100 million cells (Mumbach et al., 2016)
- Nonspecific antibodies can pull down unwanted protein complexes and contaminate the pool
- Linkers can self-ligate, generating ambiguity about true DNA interactions
- Limited sensitivity; may detect as little as 10% of interactions
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
Sati S. and Cavalli G. Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma. 2016;
References:
Ricano-Ponce I., Zhernakova D. V., Deelen P., et al. Refined mapping of autoimmune disease associated genetic variants with gene expression suggests an important role for non-coding RNAs. J Autoimmun. 2016;68:62-74
Chang H., Liu Y., Xue M., et al. Synergistic action of master transcription factors controls epithelial-to-mesenchymal transition. Nucleic Acids Res. 2016;44:2514-2527
Darabi H., Beesley J., Droit A., et al. Fine scale mapping of the 17q22 breast cancer locus using dense SNPs, genotyped within the Collaborative Oncological Gene-Environment Study (COGs). Sci Rep. 2016;6:32512
Fujimoto A., Furuta M., Totoki Y., et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48:500-509
Tewhey R., Kotliar D., Park D. S., et al. Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay. Cell. 2016;165:1519-1529
Related
History: ChIA-PET
Revision by sbrumpton on 2017-06-21 09:29:23 - Show/Hide
Chromatin Interaction Analysis by Paired-End Tag Sequencing
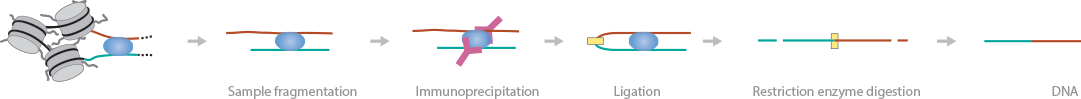
ChIA-PET features an immunoprecipitation step to map long-range DNA interactions, similar to Hi-C (Li et al., 2010) (Fullwood et al., 2009). In this method, DNA-protein complexes are crosslinked and fragmented. Specific antibodies are used to immunoprecipitate proteins of interest. Two sets of linkers, with unique barcodes, are ligated to the ends of the DNA fragments in separate aliquots, which then self-ligate based on proximity. The DNA aliquots are precipitated, digested with restriction enzymes, and sequenced. Deep sequencing provides base-pair resolution of the ligated fragments. Hi-C and ChIA-PET currently provide the best balance of resolution and reasonable coverage in the human genome to map long-range interactions (Dekker et al., 2013).
A modified protocol, called advanced or long-read ChIA-PET, has been published by Tang et al (Tang et al., 2015). This method replaces the 2 separate ligation reactions with 2 half linkers and a single biotinylated linker ligation. Next, the de-crosslinked, purified DNA is fragmented and adapters are ligated using Tn5 transposase in a single step. Finally, the DNA is PCR-amplified and sequenced (Sati et al., 2016)
Advantages:- Suitable for detecting a large number of both long-range and short-range chromatin interactions globally (Sajan et al., 2012)
- Studies the interactions made by specific proteins or protein complexes
- Public ChIA-PET datasets are available through the ENCODE Project (Consortium et al., 2011)
- Removes background generated during traditional ChIP assays
- Immunoprecipitation step reduces data complexity
Disadvantages:- Requires a large amount of starting material required, generally at least 100 million cells (Mumbach et al., 2016)
- Nonspecific antibodies can pull down unwanted protein complexes and contaminate the pool
- Linkers can self-ligate, generating ambiguity about true DNA interactions
- Limited sensitivity; may detect as little as 10% of interactions
Reagents:Illumina Library prep and Array Kit SelectorReviews:Sati S. and Cavalli G. Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma. 2016;References:Ricano-Ponce I., Zhernakova D. V., Deelen P., et al. Refined mapping of autoimmune disease associated genetic variants with gene expression suggests an important role for non-coding RNAs. J Autoimmun. 2016;68:62-74Chang H., Liu Y., Xue M., et al. Synergistic action of master transcription factors controls epithelial-to-mesenchymal transition. Nucleic Acids Res. 2016;44:2514-2527Darabi H., Beesley J., Droit A., et al. Fine scale mapping of the 17q22 breast cancer locus using dense SNPs, genotyped within the Collaborative Oncological Gene-Environment Study (COGs). Sci Rep. 2016;6:32512Fujimoto A., Furuta M., Totoki Y., et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48:500-509Tewhey R., Kotliar D., Park D. S., et al. Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay. Cell. 2016;165:1519-1529