ChIRP
Chromatin Isolation by RNA Purification
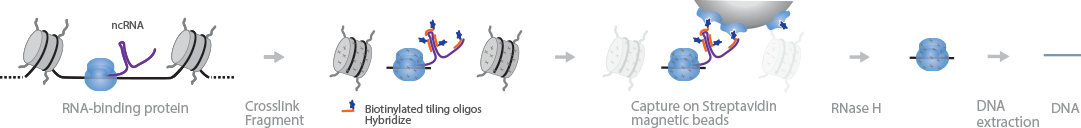
ChIRP, also commonly referred to as ChIRP-seq, is a protocol to detect the locations on the genome where ncRNAs, such as lncRNAs, and their proteins are bound (Chu et al., 2011). In this method, samples are first crosslinked and sonicated. Biotinylated tiling oligos are hybridized to the RNAs of interest, and the complexes are captured with streptavidin magnetic beads. After treatment with RNase H, the DNA is extracted and sequenced. Deep sequencing can determine the lncRNA/protein interaction site at single-base resolution.
Advantages:
- Identifies binding sites anywhere on the genome
- Enables discovery of new binding sites
- Allows selection of specific RNAs of interest
Disadvantages:
- Nonspecific oligonucleotide interactions can lead to misinterpretation of binding sites
- Chromatin can be disrupted during the preparation stage
- The sequence of the RNA of interest must be known
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
Tomita S., Abdalla M. O., Fujiwara S., et al. Roles of long noncoding RNAs in chromosome domains. Wiley Interdiscip Rev RNA. 2017;8:n/a-n/a
Schmitt A. M. and Chang H. Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452-463
Simon M. D. Insight into lncRNA biology using hybridization capture analyses. Biochim Biophys Acta. 2016;1859:121-127
Yang Y., Wen L. and Zhu H. Unveiling the hidden function of long non-coding RNA by identifying its major partner-protein. Cell Biosci. 2015;5:59
Chu C., Spitale R. C. and Chang H. Y. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:29-35
References:
Flynn R. A., Do B. T., Rubin A. J., et al. 7SK-BAF axis controls pervasive transcription at enhancers. Nat Struct Mol Biol. 2016;23:231-238
Huang W., Thomas B., Flynn R. A., et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528:517-522
Luo M., Jeong M., Sun D., et al. Long Non-Coding RNAs Control Hematopoietic Stem Cell Function. Cell Stem Cell. 2015;16:426-438
Wongtrakoongate P., Riddick G., Fucharoen S. and Felsenfeld G. Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes. PLoS Genet. 2015;11:e1005615
Li Z., Chao T. C., Chang K. Y., et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111:1002-1007
Related
History: ChIRP
Revision by sbrumpton on 2017-06-21 07:50:23 - Show/Hide
Chromatin Isolation by RNA Purification

ChIRP, also commonly referred to as ChIRP-seq, is a protocol to detect the locations on the genome where ncRNAs, such as lncRNAs, and their proteins are bound (Chu et al., 2011). In this method, samples are first crosslinked and sonicated. Biotinylated tiling oligos are hybridized to the RNAs of interest, and the complexes are captured with streptavidin magnetic beads. After treatment with RNase H, the DNA is extracted and sequenced. Deep sequencing can determine the lncRNA/protein interaction site at single-base resolution.
Advantages:- Identifies binding sites anywhere on the genome
- Enables discovery of new binding sites
- Allows selection of specific RNAs of interest
Disadvantages:- Nonspecific oligonucleotide interactions can lead to misinterpretation of binding sites
- Chromatin can be disrupted during the preparation stage
- The sequence of the RNA of interest must be known
Reagents:Illumina Library prep and Array Kit SelectorReviews:Tomita S., Abdalla M. O., Fujiwara S., et al. Roles of long noncoding RNAs in chromosome domains. Wiley Interdiscip Rev RNA. 2017;8:n/a-n/aSchmitt A. M. and Chang H. Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452-463Simon M. D. Insight into lncRNA biology using hybridization capture analyses. Biochim Biophys Acta. 2016;1859:121-127Yang Y., Wen L. and Zhu H. Unveiling the hidden function of long non-coding RNA by identifying its major partner-protein. Cell Biosci. 2015;5:59Chu C., Spitale R. C. and Chang H. Y. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:29-35References:Flynn R. A., Do B. T., Rubin A. J., et al. 7SK-BAF axis controls pervasive transcription at enhancers. Nat Struct Mol Biol. 2016;23:231-238Huang W., Thomas B., Flynn R. A., et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528:517-522Luo M., Jeong M., Sun D., et al. Long Non-Coding RNAs Control Hematopoietic Stem Cell Function. Cell Stem Cell. 2015;16:426-438Wongtrakoongate P., Riddick G., Fucharoen S. and Felsenfeld G. Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes. PLoS Genet. 2015;11:e1005615Li Z., Chao T. C., Chang K. Y., et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111:1002-1007
