4sUDRB-Seq
4-Thiouridine and 5,6-Dichlorobenzimidazole 1-b-D-Ribofuranoside Sequencing
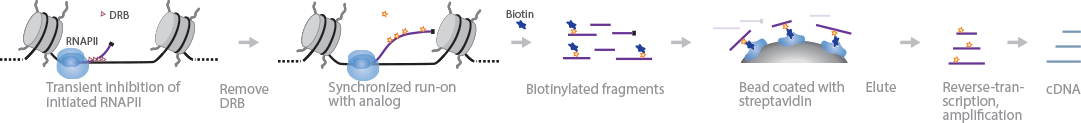
4sUDRB-Seq investigates initiation frequencies and RNA elongation rates throughout the genome using 4-thiouridine (4-SU) and DRB (Fuchs et al., 2014). Cells are first treated with DRB to inhibit RNA elongation and arrest RNAPII at TSS. The cells are lysed, cleansed of DRB, and immediately incubated with 4-SU to label newly transcribed RNA molecules. Biotin is added to bind with 4-SU, and the tagged RNA is subsequently captured using streptavidin beads. The biotinylated RNA fragments are eluted and prepared for sequencing according to the protocol in the TruSeq RNA Library Preparation Kit
Advantages:
- Measures RNA elongation rate and transcription initiation rate simultaneously
- Sequencing reads behave dynamically throughout the transcription waveãuseful in determining transcription initiation rate accurately
- No previous knowledge of the sequence is needed
Disadvantages:
- High toxicity of 4-SU can debilitate experiments with slow elongation rates
- Measuring genome-wide transition of RNAPII into active elongation can be cost-inefficient, due to the high sequencing depth needed
- Dynamic sequence reads makes identification of transcript ends challenging
- Limited to cell cultures and other artificial systems, due to the requirement for incubation in the presence of labeled nucleotides
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
Rabani M., Raychowdhury R., Jovanovic M., et al. High-Resolution Sequencing and Modeling Identifies Distinct Dynamic RNA Regulatory Strategies. Cell. 159:1698-1710
Duffy E. E., Rutenberg-Schoenberg M., Stark C. D., Kitchen R. R., Gerstein M. B. and Simon M. D. Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry. Mol Cell. 2015;59:858-866
Jonkers I. and Lis J. T. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167-177
References:
Zhang Y., Xue W., Li X., et al. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016;15:611-624
Jaenicke L. A., von Eyss B., Carstensen A., et al. Ubiquitin-Dependent Turnover of MYC Antagonizes MYC/PAF1C Complex Accumulation to Drive Transcriptional Elongation. Mol Cell. 2016;61:54-67
Fuchs G., Rosenthal E., Bublik D.-R., Kaplan T. and Oren M. Gene body H2B monoubiquitylation regulates gene-selective RNA Polymerase II pause release and is not rate limiting for transcription elongation. bioRxiv. 2015;
Related
History: 4sUDRB-Seq
Revision by sbrumpton on 2017-06-21 09:06:28 - Show/Hide
4-Thiouridine and 5,6-Dichlorobenzimidazole 1-b-D-Ribofuranoside Sequencing

4sUDRB-Seq investigates initiation frequencies and RNA elongation rates throughout the genome using 4-thiouridine (4-SU) and DRB (Fuchs et al., 2014). Cells are first treated with DRB to inhibit RNA elongation and arrest RNAPII at TSS. The cells are lysed, cleansed of DRB, and immediately incubated with 4-SU to label newly transcribed RNA molecules. Biotin is added to bind with 4-SU, and the tagged RNA is subsequently captured using streptavidin beads. The biotinylated RNA fragments are eluted and prepared for sequencing according to the protocol in the TruSeq RNA Library Preparation Kit
Advantages:- Measures RNA elongation rate and transcription initiation rate simultaneously
- Sequencing reads behave dynamically throughout the transcription waveãuseful in determining transcription initiation rate accurately
- No previous knowledge of the sequence is needed
Disadvantages:- High toxicity of 4-SU can debilitate experiments with slow elongation rates
- Measuring genome-wide transition of RNAPII into active elongation can be cost-inefficient, due to the high sequencing depth needed
- Dynamic sequence reads makes identification of transcript ends challenging
- Limited to cell cultures and other artificial systems, due to the requirement for incubation in the presence of labeled nucleotides
Reagents:Illumina Library prep and Array Kit SelectorReviews:Rabani M., Raychowdhury R., Jovanovic M., et al. High-Resolution Sequencing and Modeling Identifies Distinct Dynamic RNA Regulatory Strategies. Cell. 159:1698-1710Duffy E. E., Rutenberg-Schoenberg M., Stark C. D., Kitchen R. R., Gerstein M. B. and Simon M. D. Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry. Mol Cell. 2015;59:858-866Jonkers I. and Lis J. T. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167-177References:Zhang Y., Xue W., Li X., et al. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016;15:611-624Jaenicke L. A., von Eyss B., Carstensen A., et al. Ubiquitin-Dependent Turnover of MYC Antagonizes MYC/PAF1C Complex Accumulation to Drive Transcriptional Elongation. Mol Cell. 2016;61:54-67Fuchs G., Rosenthal E., Bublik D.-R., Kaplan T. and Oren M. Gene body H2B monoubiquitylation regulates gene-selective RNA Polymerase II pause release and is not rate limiting for transcription elongation. bioRxiv. 2015;
