SHAPE-Seq
Selective 2ê-Hydroxyl Acylation Analyzed by Primer Extension Sequencing
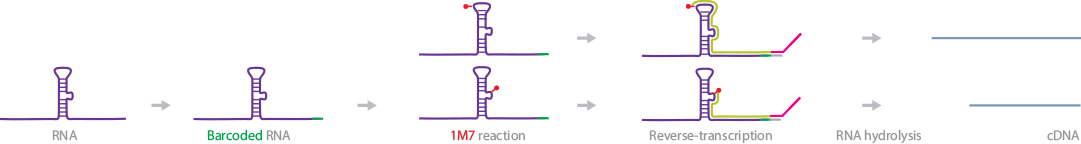
SHAPE-Seq provides structural information about RNA (Lucks et al., 2011) (Watters et al., 2016). In this method, a unique barcode is first added to the 3′ end of RNA, and the RNA is allowed to fold under pre-established in vitro conditions. The barcoded and folded RNA is treated with a SHAPE reagent, 1-methyl-7-nitroisatoic anhydride (1M7), which blocks RT. The RNA is reverse-transcribed to cDNA. Deep sequencing of the cDNA provides single-nucleotide sequence information for the positions occupied by 1M7. The structural information of the RNA can then be deduced.
Advantages:
- Provides RNA structural information
- Multiplexed analysis of barcoded RNAs provides information for multiple RNAs
- Effect of point mutations on RNA structure can be assessed
- Alternative to mass spectrometry, nuclear magnetic resonance (NMR), and crystallography
Disadvantages:
- Requires positive and negative controls to account for transcriptase drop-off
- Requires pre-established conditions for RNA folding
- Folding in vitro may not reflect actual folding in vivo
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
Fang Y. and Fullwood M. J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics Proteomics Bioinformatics. 2016;14:42-54
References:
Taliaferro J. M., Lambert N. J., Sudmant P. H., Dominguez D., Merkin J. J., et al. RNA Sequence Context Effects Measured In Vitro Predict In Vivo Protein Binding and Regulation. Mol Cell. 2016;64:294-306
Watters K. E., Abbott T. R. and Lucks J. B. Simultaneous characterization of cellular RNA structure and function with in-cell SHAPE-Seq. Nucleic Acids Res. 2015;
Related
History: SHAPE-Seq
Revision by sbrumpton on 2017-06-21 09:06:28 - Show/Hide
Selective 2ê-Hydroxyl Acylation Analyzed by Primer Extension Sequencing

SHAPE-Seq provides structural information about RNA (Lucks et al., 2011) (Watters et al., 2016). In this method, a unique barcode is first added to the 3' end of RNA, and the RNA is allowed to fold under pre-established in vitro conditions. The barcoded and folded RNA is treated with a SHAPE reagent, 1-methyl-7-nitroisatoic anhydride (1M7), which blocks RT. The RNA is reverse-transcribed to cDNA. Deep sequencing of the cDNA provides single-nucleotide sequence information for the positions occupied by 1M7. The structural information of the RNA can then be deduced.
Advantages:- Provides RNA structural information
- Multiplexed analysis of barcoded RNAs provides information for multiple RNAs
- Effect of point mutations on RNA structure can be assessed
- Alternative to mass spectrometry, nuclear magnetic resonance (NMR), and crystallography
Disadvantages:- Requires positive and negative controls to account for transcriptase drop-off
- Requires pre-established conditions for RNA folding
- Folding in vitro may not reflect actual folding in vivo
Reagents:Illumina Library prep and Array Kit SelectorReviews:Fang Y. and Fullwood M. J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics Proteomics Bioinformatics. 2016;14:42-54References:Taliaferro J. M., Lambert N. J., Sudmant P. H., Dominguez D., Merkin J. J., et al. RNA Sequence Context Effects Measured In Vitro Predict In Vivo Protein Binding and Regulation. Mol Cell. 2016;64:294-306Watters K. E., Abbott T. R. and Lucks J. B. Simultaneous characterization of cellular RNA structure and function with in-cell SHAPE-Seq. Nucleic Acids Res. 2015;
