hiCLIP
RNA Hybrid and Individual-Nucleotide Resolution Ultraviolet Crosslinking and Immunoprecipitation
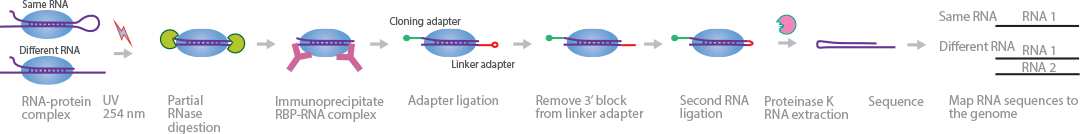
hiCLIP sequences RNA duplexes bound to RBPs in vivo (Sugimoto et al., 2015). The unique cloning and linker-adapter system in hiCLIP identifies whether the RBP-bound duplex originates from the same RNA or different RNAs.
Similar to CLIP library preparation techniques, RNA and RBPs are UV-crosslinked, partially digested, and immunoprecipitated. The signature hiCLIP cloning and linker-adapters are ligated to both strands of the duplex. The 3′ end of the linker-adapter is dephosphorylated and ligated to the 5′ end of the other strand. The bound proteins are removed with proteinase K. The cDNA library is prepared in a similar dashion to iCLIP for high-throughput sequencing. The cloning and linker-adapters enable the mapping of the hybrid reads to the transcriptome and distinguish whether the duplex is formed by the same RNAs or different RNAs.
Advantages:
- Maps RBP-bound RNA duplexes in vivo
- Linker-adapter remediates physical and computational challenges seen in crosslinking, ligation, and sequencing of hybrids (CLASH)
- Detects long-range RNA duplex interactions (Lu et al., 2016)
Disadvantages:
- Does not identify non-RBP-bound RNA duplexes
- Technically complex procedure
- Uses radioactive labeling
- Artifacts may be introduced in the circularization step
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
Lu Z. and Chang H. Y. Decoding the RNA structurome. Curr Opin Struct Biol. 2016;36:142-148
Weidmann C. A., Mustoe A. M. and Weeks K. M. Direct Duplex Detection: An Emerging Tool in the RNA Structure Analysis Toolbox. Trends Biochem Sci. 2016;41:734-736
Nawy T. Structural biology: RNA structure served in vivo. Nature Methods. 2015;12:383-383
Burgess D. J. RNA. Detailed probing of RNA structure in vivo. Nat Rev Genet. 2015;16:255
References:
Sugimoto Y., Vigilante A., Darbo E., et al. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015;519:491-494
Related
History: hiCLIP
Revision by sbrumpton on 2017-06-21 07:50:24 - Show/Hide
RNA Hybrid and Individual-Nucleotide Resolution Ultraviolet Crosslinking and Immunoprecipitation

hiCLIP sequences RNA duplexes bound to RBPs in vivo (Sugimoto et al., 2015). The unique cloning and linker-adapter system in hiCLIP identifies whether the RBP-bound duplex originates from the same RNA or different RNAs.
Similar to CLIP library preparation techniques, RNA and RBPs are UV-crosslinked, partially digested, and immunoprecipitated. The signature hiCLIP cloning and linker-adapters are ligated to both strands of the duplex. The 3' end of the linker-adapter is dephosphorylated and ligated to the 5' end of the other strand. The bound proteins are removed with proteinase K. The cDNA library is prepared in a similar dashion to iCLIP for high-throughput sequencing. The cloning and linker-adapters enable the mapping of the hybrid reads to the transcriptome and distinguish whether the duplex is formed by the same RNAs or different RNAs.
Advantages:- Maps RBP-bound RNA duplexes in vivo
- Linker-adapter remediates physical and computational challenges seen in crosslinking, ligation, and sequencing of hybrids (CLASH)
- Detects long-range RNA duplex interactions (Lu et al., 2016)
Disadvantages:- Does not identify non-RBP-bound RNA duplexes
- Technically complex procedure
- Uses radioactive labeling
- Artifacts may be introduced in the circularization step
Reagents:Illumina Library prep and Array Kit SelectorReviews:Lu Z. and Chang H. Y. Decoding the RNA structurome. Curr Opin Struct Biol. 2016;36:142-148Weidmann C. A., Mustoe A. M. and Weeks K. M. Direct Duplex Detection: An Emerging Tool in the RNA Structure Analysis Toolbox. Trends Biochem Sci. 2016;41:734-736Nawy T. Structural biology: RNA structure served in vivo. Nature Methods. 2015;12:383-383Burgess D. J. RNA. Detailed probing of RNA structure in vivo. Nat Rev Genet. 2015;16:255References:Sugimoto Y., Vigilante A., Darbo E., et al. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015;519:491-494
