TIVA
Transcriptome in Vivo Analysis
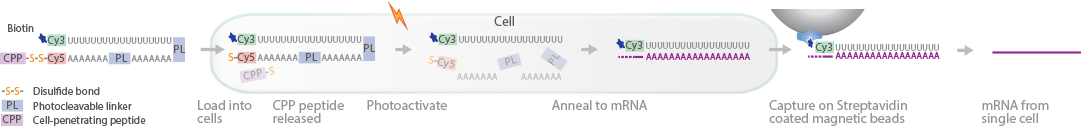
TIVA is a protocol that captures mRNA from live cells (Lovatt et al., 2014). In this method, a photoactivatable TIVA tag is loaded into cells. Selective photoactivation exposes the mRNA-capturing portion of the tag, allowing it to hybridize to the poly(A) tails of mRNA. The biotin-bound mRNA is captured using streptavidin-coated magnetic beads and transcribed into cDNA. Sequencing the cDNA provides transcriptome analysis of RNA from single cells in complex tissues.
Advantages:
- in vivo transcriptome analysis using photoactivation in neuronal cells
- Noninvasive method for capturing mRNAs in their natural microenvironment
Disadvantages:
- Limited to a small number of cells
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
Zhang X., Marjani S. L., Hu Z., Weissman S. M., Pan X., et al. Single-Cell Sequencing for Precise Cancer Research: Progress and Prospects. Cancer Res. 2016;76:1305-1312
Binan L., Mazzaferri J., Choquet K., et al. Live single-cell laser tag. Nat Commun. 2016;7:11636
Bian Q. and Cahan P. Computational Tools for Stem Cell Biology. Trends Biotechnol. 2016;
Liu S. and Trapnell C. Single-cell transcriptome sequencing: recent advances and remaining challenges. F1000Res. 2016;5:
Achim K., Pettit J. B., Saraiva L. R., et al. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat Biotechnol. 2015;
Kolodziejczyk A. A., Kim J. K., Svensson V., Marioni J. C. and Teichmann S. A. The Technology and Biology of Single-Cell RNA Sequencing. Mol Cell. 2015;58:610-620
Baslan T. and Hicks J. Single cell sequencing approaches for complex biological systems. Curr Opin Genet Dev. 2014;26C:59-65
References:
Lovatt D., Ruble B. K., Lee J., et al. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat Methods. 2014;11:190-196
Related
History: TIVA
Revision by sbrumpton on 2017-06-21 07:50:22 - Show/Hide
Transcriptome in Vivo Analysis

TIVA is a protocol that captures mRNA from live cells (Lovatt et al., 2014). In this method, a photoactivatable TIVA tag is loaded into cells. Selective photoactivation exposes the mRNA-capturing portion of the tag, allowing it to hybridize to the poly(A) tails of mRNA. The biotin-bound mRNA is captured using streptavidin-coated magnetic beads and transcribed into cDNA. Sequencing the cDNA provides transcriptome analysis of RNA from single cells in complex tissues.
Advantages:- in vivo transcriptome analysis using photoactivation in neuronal cells
- Noninvasive method for capturing mRNAs in their natural microenvironment
Disadvantages:- Limited to a small number of cells
Reagents:Illumina Library prep and Array Kit SelectorReviews:Zhang X., Marjani S. L., Hu Z., Weissman S. M., Pan X., et al. Single-Cell Sequencing for Precise Cancer Research: Progress and Prospects. Cancer Res. 2016;76:1305-1312Binan L., Mazzaferri J., Choquet K., et al. Live single-cell laser tag. Nat Commun. 2016;7:11636Bian Q. and Cahan P. Computational Tools for Stem Cell Biology. Trends Biotechnol. 2016;Liu S. and Trapnell C. Single-cell transcriptome sequencing: recent advances and remaining challenges. F1000Res. 2016;5:Achim K., Pettit J. B., Saraiva L. R., et al. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat Biotechnol. 2015;Kolodziejczyk A. A., Kim J. K., Svensson V., Marioni J. C. and Teichmann S. A. The Technology and Biology of Single-Cell RNA Sequencing. Mol Cell. 2015;58:610-620Baslan T. and Hicks J. Single cell sequencing approaches for complex biological systems. Curr Opin Genet Dev. 2014;26C:59-65References:Lovatt D., Ruble B. K., Lee J., et al. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat Methods. 2014;11:190-196
