TCR Chain Pairing
Identification of T-Cell Receptor (TCR) a-b Chain Pairing in Single Cells
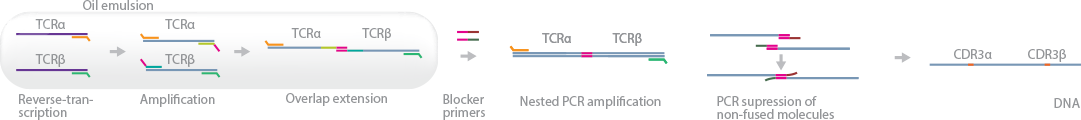
This method identifies TCR-alpha and -beta chain pairing in single cells using cell-based emulsion technology for isolation, followed by NGS (Turchaninova et al., 2013). TCR chain pairing resolves one of the biggest challenges of identifying coexpressed gene pairsãrandom, nonspecific overlap extension of nonfused moleculesãby introducing a unique PCR suppression technique during post_emulsion amplification reactions.
First, single T cells are isolated into oil emulsion droplets containing RT primers for _ and _ chain mRNA strands. The resultant cDNA is amplified, and the RT primer extensions overlapped to connect TCR-_ and TCR-_ strands, which are now called fused molecules. The cDNA products are extracted from the emulsion and introduced to blocking primers. These primers anneal to the 3′ end of nonfused cDNA strands, preventing them from being amplified; the authors name this technique –PCR suppression.” Finally, the fused molecules are PCR-amplified and sequenced.
Similar methods: TCR-LA-MC PCR
Advantages:
- Identifies TCR-alpha-beta chain pairing using NGS
- PCR suppression of nonfused molecules significantly reduces random, nonspecific overlap extension during post_emulsion amplification reactions
- Simple and straightforward protocol
Disadvantages:
- Heat-shock during cell lysis may reduce enzymatic activity during RT (Friedensohn et al., 2017)
- Amplification product is not suitable for cloning (Sprouse et al., 2016)
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
Friedensohn S., Khan T. A. and Reddy S. T. Advanced Methodologies in High-Throughput Sequencing of Immune Repertoires. Trends in Biotechnology. 2017;35:203-214
References:
Munson D. J., Egelston C. A., Chiotti K. E., et al. Identification of shared TCR sequences from T cells in human breast cancer using emulsion RT-PCR. Proc Natl Acad Sci U S A. 2016;113:8272-8277