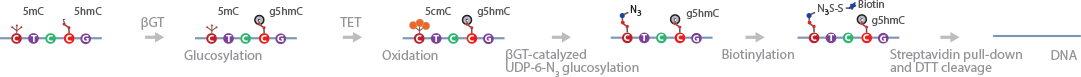TAmC-Seq
Tet-Assisted 5-Methylcytosine Sequencing
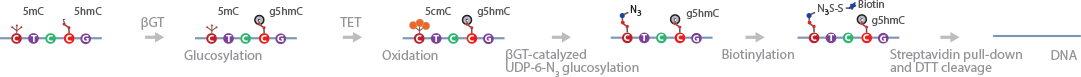
TAmC-Seq selectively tags 5mC with an azide residue that can be further labeled with biotin for affinity purification (Zhang et al., 2013). The method relies on the properties of the Ten-eleven translocation (Tet) family of iron(II)/α-ketoglutarate-dependent dioxygenases that are involved in the demethylation of 5mC.
In this method, the endogenous 5hmC in genomic DNA is protected by glucosylation with normal glucose. The 5mC residues are converted to 5hmC via mTet1-catalyzed oxidation. The 5hmC is then labeled via β-GT-mediated glucosylation with a modified glucose moiety (6-N3-glucose) to generate 6-N3-β-glucosyl-5-hydroxymethylcytosine (N3-5gmC). This molecule can be labeled further, using click chemistry, for detection, affinity purification, and sequencing.
Advantages:
- Captures a larger fraction of methylated CpGs with less density bias
Disadvantages:
- Not yet adopted widely by the scientific community
Reagents:
Illumina Library prep and Array Kit Selector
Reviews:
None available yet
References:
Zhang L., Szulwach K. E., Hon G. C., et al. Tet-mediated covalent labelling of 5-methylcytosine for its genome-wide detection and sequencing. Nat Commun. 2013;4:1517
Related
History: TAmC-Seq
Revision by sbrumpton on 2017-06-21 07:50:20 - Show/Hide
Tet-Assisted 5-Methylcytosine Sequencing
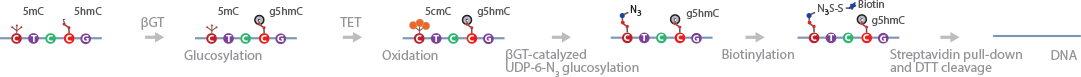
TAmC-Seq selectively tags 5mC with an azide residue that can be further labeled with biotin for affinity purification (Zhang et al., 2013). The method relies on the properties of the Ten-eleven translocation (Tet) family of iron(II)/α-ketoglutarate-dependent dioxygenases that are involved in the demethylation of 5mC.
In this method, the endogenous 5hmC in genomic DNA is protected by glucosylation with normal glucose. The 5mC residues are converted to 5hmC via mTet1-catalyzed oxidation. The 5hmC is then labeled via β-GT-mediated glucosylation with a modified glucose moiety (6-N3-glucose) to generate 6-N3-β-glucosyl-5-hydroxymethylcytosine (N3-5gmC). This molecule can be labeled further, using click chemistry, for detection, affinity purification, and sequencing.
Advantages:- Captures a larger fraction of methylated CpGs with less density bias
Disadvantages:- Not yet adopted widely by the scientific community
Reagents:Illumina Library prep and Array Kit SelectorReviews:None available yet
References:Zhang L., Szulwach K. E., Hon G. C., et al. Tet-mediated covalent labelling of 5-methylcytosine for its genome-wide detection and sequencing. Nat Commun. 2013;4:1517