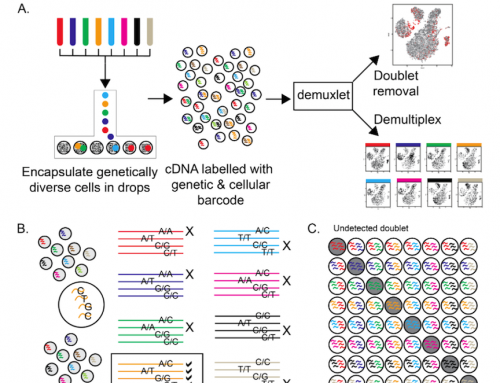
The use of DNA methylation analysis by NGS has become a standard tool in many labs. In a project design discussion we had today somebody mentioned the use of a control for bisulfite conversion efficiency that I’d missed, as its such a simple one I thought I’d briefly mention it here. In their PLoS Genet 2013 paper, Shirane et al from Kyushu University spiked-in unmethylated lambda phage DNA (Promega) to control for, and check, the C/T conversion rate was greater than 99%.
The bisulfite conversion of cytosine bases to uracils, by deamination of unmethylated cytosine (as shown above) is the gold standard for methylation analysis.
Users identify the C/T transitions in a comparison of bisufite treated/untreated samples, or by comparing to a known reference. However bisulfite treatment is a harsh biochemical reaction, and can cause large losses in template DNA. As such controlling for and measuring conversion efficiency is important in making conclusions about the methylation data from NGS experiments. As a reminder – Bisulfite does not convert methylated or hydroxy-mehtylated cytosine allowing users to discriminate between non-methylcytosine (C) and methylcytosine (mC) or hydroxymethylated (hmC) cytosine.
Users identify the C/T transitions in a comparison of bisufite treated/untreated samples, or by comparing to a known reference. However bisulfite treatment is a harsh biochemical reaction, and can cause large losses in template DNA. As such controlling for and measuring conversion efficiency is important in making conclusions about the methylation data from NGS experiments. As a reminder – Bisulfite does not convert methylated or hydroxy-mehtylated cytosine allowing users to discriminate between non-methylcytosine (C) and methylcytosine (mC) or hydroxymethylated (hmC) cytosine.
We’re likely to start using this control if it works well in the project we have just kicked off. In the paper they added 1ng of to 1000 oocytes before performing a PBAT analysis. We’ll aim for 1% spike-in, but need to consider how much to add to each sample, and whether Lambda is the right spike-in as we’re using an RRBS method or this project. To check the suitability I grabbed the Lambda sequence from Genbank and did an in silico Msp1 digest using WebCutter2.0. I found 330 cut sites – which should be plenty for checking efficiency.
Want to learn more about bisulfite conversion in general? Take a look at Zymo‘s website, it’s an excellent resource.




Very interesting and appreciate the resource.